上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
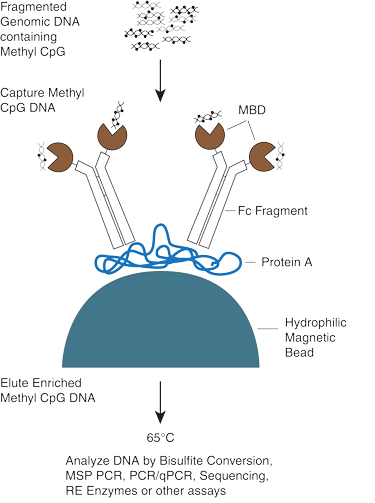
此试剂盒通过人 MBD2 蛋白的甲基化 CpG 结合域将甲基化 DNA 从片段化基因组 DNA (5 ng–25 μg) 中分离出来,其中,人 MBD2 蛋白融合了人 IgG1 抗体的 Fc 尾部(MBD2-Fc)并偶联顺磁性 Protein A 磁珠(MBC2-Fc/Protein A 磁珠)。Protein A 上的一个位点可高亲和力地结合两个 Fc 结构域(Kd=10-7)。由于 Fc 片段是二聚体,因此在溶剂中,每个 Protein A 分子有四个暴露的 MBD2 结构域结合位点,从而将相对平衡常数提高了 100 倍。这种稳定的复合物可以选择性地结合含甲基化 CpG 的双链 DNA。经过简单漂洗和磁性捕获步骤之后,富集的 DNA 样本可以在 65℃ 下很容易地在少量无核酸酶水中温育洗脱,并且可随即对样本进行下游分析。下游分析方法有多种,包括:
- 终点和实时 PCR 测定
- 重亚硫酸盐转化后进行 DNA 扩增
- 克隆和测序
- 直接测序
- 高通量测序文库制备
- 针对 DNA 微阵列分析进行标记
- 基于甲基化敏感限制性内切酶的测定
MBD-Fc 和 MCIp 最初由雷根斯堡大学的 Michael Rehli 开发,用于提高传统 CpG 结合技术的敏感性和特异性。
试剂盒组分:
每个试剂盒包含的试剂足以从最高 100 μg 片段化的起始 DNA 中富集甲基化 DNA。如果每次实验的 DNA 起始样本量为 5 ng 到 10 μg,则此试剂盒提供的试剂足以用于 25 次反应。贮存温度为 4℃。如果存放时间超过 6 个月,则应将 MBD2-Fc 蛋白存放在 -20℃ 温度以下。
- 产品类别:
- Methylome Analysis Products,
- Epigenetic Analysis Products,
- Next Generation Sequencing Library Preparation Products
- 应用:
- DNA Enrichment
-
试剂盒组成
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E2600S Multi-temperature Bind/Wash Reaction Buffer (5X) B1032AVIAL 4 2 x 25 ml 5 X NaCl High-Salt Elution Buffer (2M) B1033AVIAL 4 1 x 15 ml Not Applicable Fragmented HeLa DNA N0484AVIAL 4 1 x 0.02 ml 100 µg/ml LINE Primers for Methylated Controls N0485AVIAL 4 1 x 0.02 ml 100 µM RPL30 Primers for Non-methylated Controls N0486AVIAL 4 1 x 0.02 ml 100 µM MirA Primers for Input Control N0487AVIAL 4 1 x 0.02 ml 100 µM MBD2a-Fc Protein M0358AVIAL -20 1 x 0.025 ml 2 mg/ml Protein A Magnetic Beads S1426AVIAL 4 1 x 0.25 ml Not Applicable
-
-
特性和用法
需要但不提供的材料
50 bp DNA Ladder(NEB #N3236)
6 孔磁性分离架(NEB #S1506)
Taq DNA 聚合酶(提供标准 Taq 缓冲液)(NEB #M0273)
脱氧核糖核苷酸混合溶液(NEB #N0447)
无核酸酶水 -
优势和特性
Features
- 高亲和性结合力带来更高灵敏度。
- 减少洗脱液用量可简化下游实验。
- 操作简便,样本富集时间不到 2 小时。
- 已富集的甲基化 DNA 样本可轻松连接双链接头,以便进行二代测序。
- 从不同浓度范围的 DNA 投入量可获得纯度高的产物。
-
方法概述
方法概述
第一步 — 片段化基因组 DNA
必须采用超声处理、雾化或酶处理方式将 DNA 片段化为平均长度小于 1,000 bp。第二步 — 在 1X 结合/漂洗缓冲液中使 MBD2-Fc 与 Protein A 磁珠结合。
在室温下温育反应 15 分钟。在结合/漂洗反应缓冲液中漂洗磁珠两次。第三步 — 将片段化的 DNA 添加到 MBD2-Fc/Protein A 磁珠里。
在室温下温育反应 20 分钟。在室温下漂洗磁珠三次,每次漂洗五分钟,去掉未结合的 DNA。第四步 — 将富集的甲基化 CpG DNA 从磁珠上洗脱
在 65℃ 温度下,在无 DNase 的水中温育样本 15 分钟。
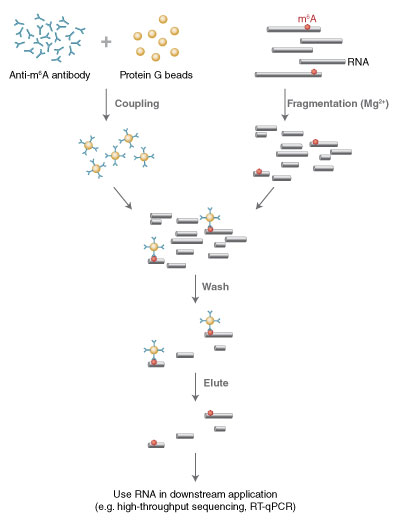
图 1:富集流程
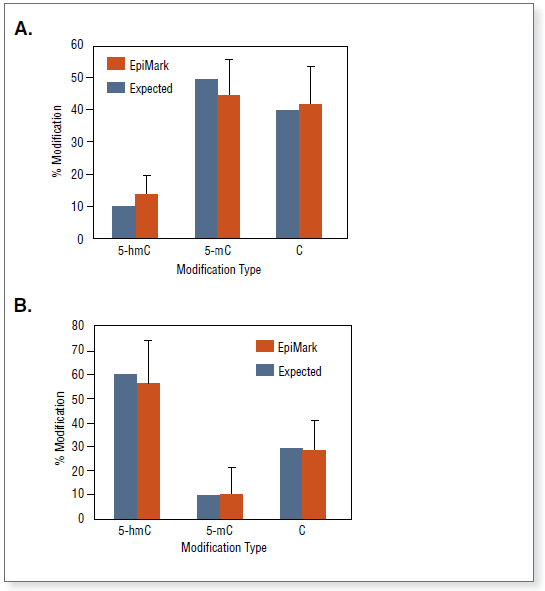
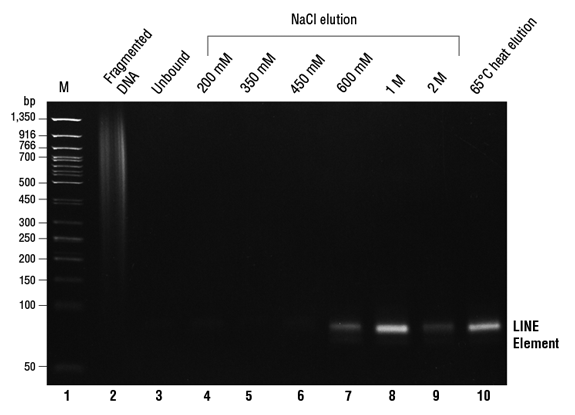
图 2:对使用 EpiMark 甲基化 DNA 富集试剂盒纯化的样本进行甲基化位点的终点 PCR 分析(LINE)。
已分析的样本显示在胶图上方。
-
相关产品
相关产品
- Taq DNA 聚合酶(提供标准 Taq 缓冲液)
- 50 bp DNA Ladder
- 6 孔磁性分离架
- Deoxynucleotide Solution Mix
-
参考文献
- Cross, S.H., Charlton, J.A., Nan, X., and Bird, A.P. (1994). Nat. Genet. . 6, 236-244.
- Fraga, M.F. et al. (2003). Nucleic Acids Res.. 31, 1765-1774.
- Gebhard, C. et al. (2006). Nucleic Acids Res. . 34, e82.
- Gebhard, C. et al. (2006). Cancer Research . 66, 6118-6128.
- Jacinto, F.V., Ballestar, E. and Esteller, M. (2008). Biotechniques . 44, 35, 37, 39 passim.
- Lister, R. et al. (2008). Cell. 133, 523-536.
- Rauch, T. and Pfeifer, G.P. (2005). Lab Invest.. 85, 1172-1180.
- Schilling, E. and Rehli, M. (2007). Genomics . 90, 314-323.
- Weber, M. et al. (2005). Nat. Genet. . 37, 853-862.
操作说明、说明书 & 用法
-
操作说明
- DNA Fragmentation (E2600)
- Prebind MBD2a-Fc to Protein A Magnetic Beads (NEB #E2600)
- Capture Methylated CpG DNA (E2600)
- Wash Off Unbound DNA (E2600)
- Elute Captured Methylated CpG DNA (E2600)
- Capture and Elute Step for Control DNA (E2600)
- Downstream Analysis (NEB #E2600)
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE2600
FAQs & 问题解决指南
-
问题解决指南
- EpiMark® Methylated DNA Enrichment Kit Troubleshooting Guide