上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
In the NEBNext Direct target enrichment approach (Figure 1), enzymatically nicked or Covaris® sheared DNA is hybridized to biotinylated oligonucleotide baits that capture both strands of the target DNA and define the 3´ ends of the regions of interest. After hybridization, the bait-target hybrids are bound to streptavidin beads and any 3´ off-target sequence is removed enzymatically. This combination of hybridization with enzymatic removal of 3´ off-target sequence enables greater sequencing specificity relative to conventional hybridization-based enrichment methods. The trimmed targets are then converted into Illumina-compatible libraries that include a 12 bp unique molecular identifier (UMI) in the Illumina i5 index location and an 8 bp sample barcode in the Illumina i7 index location. The NEBNext Direct enrichment method can be performed within one to two days and is compatible with most automated liquid handling instruments.
The NEBNext Direct Cancer HotSpot Panel is designed to enrich for DNA fragments across 190 common cancer targets from 50 genes, encompassing approximately 40 kb of sequence and including over 18,000 COSMIC features. This kit contains the oligonucleotides, beads, enzymes and buffers required to convert the desired fragments into a sequence-ready library for next-generation sequencing on the Illumina platform and is designed for PE75 or PE150 Illumina sequencing.
Advantages
- Generate a higher percentage of your sequencing reads aligning to your targets
- Eliminate the need to over-sequence, reducing cost per sample
- Obtain uniform sequencing of all targets, regardless of GC content
- Save time with a 1-day workflow that combines enrichment with library preparation
- Generate high quality libraries with limited input amounts and degraded DNA samples, including FFPE and ctDNA
- Distinguish molecular duplicates, reducing false positive variants and improving sensitivity
Figure 1. NEBNext Direct employs a fast hybridization-based workflow that combines capture with library preparation.
Table 1. Targets include regions from the following cancer-related genes:
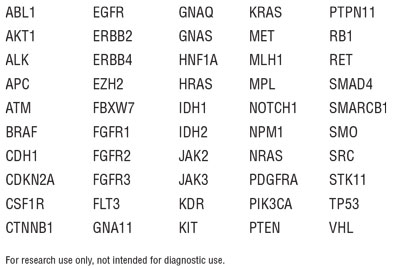
Figure 2. The NEBNext Direct Cancer HotSpot Panel demonstrates the ability to accurately detect a range of nucleic acid variants.
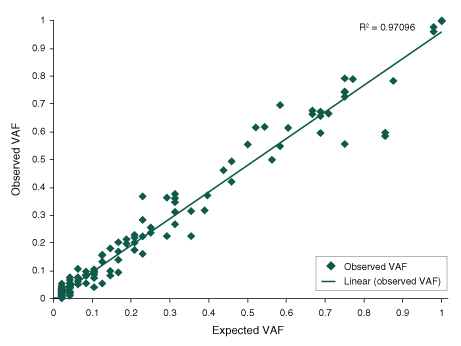
Figure 3. The NEBNext Direct Cancer HotSpot Panel delivers a high percentage of sequence reads mapping to targets, even with challenging sample types.
- Graph shows the percentage of aligned sequence reads that map to the targets
- 100 ng of DNA was used for each library preparation
- Reads were generated on an Illumina® MiSeq® with 2 x 75 bp reads, 8 bp sample ID, and 12 bp unique molecule ID
- Alignments were performed with BWA-MEM and PCR duplicates were filtered using the unique molecule IDs
Figure 4. The NEBNext Direct Cancer HotSpot Panel displays high uniformity of coverage across targets.
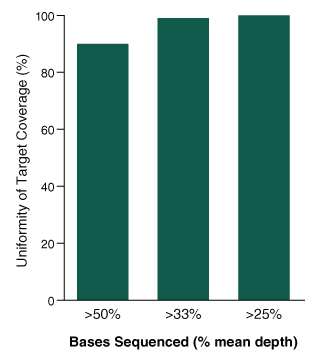
- Graph shows the percentage of target bases sequenced to at least 50%, 33%, and 25% of the mean read depth
- 100 ng of DNA was used for each library preparation
- Reads were generated on an Illumina MiSeq with 2 x 75 bp reads, 8 bp sample ID, and 12 bp unique molecule ID
- Alignments were performed with BWA-MEM and PCR duplicates were filtered using the unique molecule IDs
Figure 5. The NEBNext Direct Cancer HotSpot Panel offers minimized bias across sequence content.
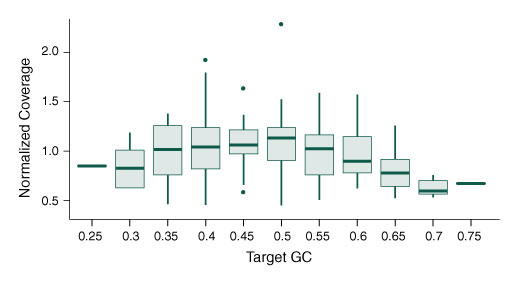
- Graph shows the normalized depth of coverage of targets of varying GC content
- 100 ng of DNA was used for each library preparation
- Reads were generated on an Illumina MiSeq with 2 x 75 bp reads, 8 bp sample ID, and 12 bp unique molecule ID
- Alignments were performed with BWA-MEM and PCR duplicates were filtered using the unique molecule IDs
ILLUMINA® and MISEQ® are registered trademarks of Illumina®, Inc.
批次质控
所提供的单独批次均通过额外功能验证。单个试剂通过标准酶活性和质控分析,并同时满足各单独组分页面列出的严格的附加质控标准
- 产品类别:
- Discontinued (<3 years)
-
特性和用法
需要但不提供的材料
- Covaris® microTubes or plate
- 1X TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA)
- Molecular grade ethanol
- Molecular grade water
- 96-well PCR plates (or PCR strip tubes)
- Eppendorf® DNA LoBind® 2 ml tubes (VWR, cat#: 80077-234)
- Additional microcentrifuge or conical tubes to prepare master mixes
- 96-well plate magnet or PCR tube magnet
- Microcentrifuge tube magnet
- Agilent® High Sensitivity DNA Kit (Agilent, cat#: 5067-4626)
Required Equipment:- Covaris Focused-Ultrasonicator
- Thermocycler programmable to 100 μl
- Agilent Bioanalyzer® or similar instrument
操作说明、说明书 & 用法
-
操作说明
- Guidelines for Setting Up PCR Reactions (E7000X only)
- Protocol for NEBNext Direct® Cancer HotSpot Panel (NEB #E7000)
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE7000
-
使用指南
- GRCh37hg19 BED files for the NEBNext Direct Cancer HotSpot Panel
- GRCh38/hg38 BED files for the NEBNext Direct® Cancer HotSpot Panel
- Index Pooling Guidelines for E7000
- Sample Sheet for E7000L with the index sequences that can be used in the Illumina® experiment Manager v4
- Sample Sheet for E7000L with the index sequences that can be used in the Illumina® experiment Manager v5
- Sample Sheet for E7000S with the index sequences that can be used in the Illumina® experiment Manager v4
- Sample Sheet for E7000S with the index sequences that can be used in the Illumina® experiment Manager v5
- Sample Sheet for E7000X with the index sequences that can be used in the Illumina® experiment Manager v4
- Sample Sheet for E7000X with the index sequences that can be used in the Illumina® experiment Manager v5
- Using Unique Molecular IDs with NEBNext Direct® – Data Usage Guideline Page
FAQs & 问题解决指南
-
FAQs
- Can NEBNext Direct be applied for genotyping applications?
- What size should I fragment my DNA to?
- If my FFPE DNA is fragmented, should I do a shearing step?
- Can I repair my FFPE DNA using the NEBNext FFPE DNA Repair Mix (NEB #M6630)?
- How can I warm Bead Wash 1 (BW1) to dissolve any precipitate?
- If I have adaptor dimer in my finished library, can I perform another bead cleanup to remove it?
- How many times can the -20°C reagents be frozen and thawed?
- How many times can the 4°C reagents be brought to room temperature?
- My 4°C box accidentally froze, can I still use it?
- I accidentally stored the Sample Purification Beads and Bead Wash 1 (BW1) at 4°C. Can I still use them?
- What method do you recommend for extracting genomic DNA and FFPE DNA before use with the NEBNext Direct Panel?
- Can shearing be done using Covaris®?
- What is the recommended method for quantitating my input DNA?
- If my DNA input material amount is high, should I reduce the number of PCR cycles?
- If my input amount is low, should I dilute the adaptors?
- What type and how much starting material do I need to use when preparing libraries using the NEBNext Direct Panel?
- Which magnets (2 ml and 200 µl sizes) do you recommend?
- Is it ok to leave bead separations for longer than 15 seconds?
- I used Bead Wash 1 (BW1) when I should have used Bead Wash 2 (BW2) during the Post-reaction Wash. Can I do another wash with Bead Wash 2 (BW2) to save my sample?
- I used Bead Wash 2 (BW2) when I should have used Bead Wash 1 (BW1) during the Post-reaction Wash. What should I do?
- Does incomplete removal of Bead Wash 1 prior to adding Bead Wash 2 inhibit the next step?
- Can I use any Q5® formulation with this kit?
- How many sequencing reads per sample do you recommend using with the NEBNext Direct Cancer HotSpot Panel?
-
How many/ few samples can I pool together for my sequencing run?
- Are the indexes in E7000S/ E6627S/E6631S (D01-D08) the same sequences as the first 8 indexes (D01-D08) in NEB #E7000L/ E6627L/E6631L?
- What is the shelf life of this product?
