上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
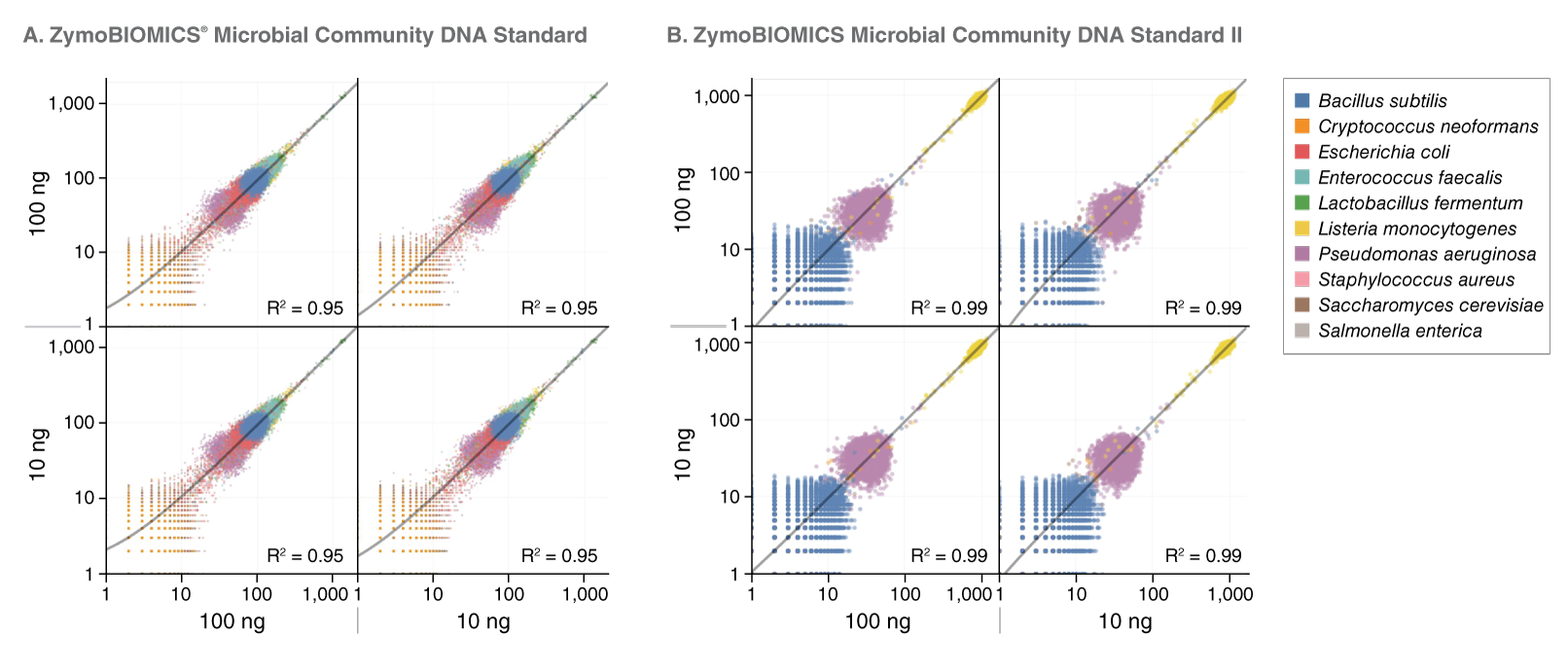
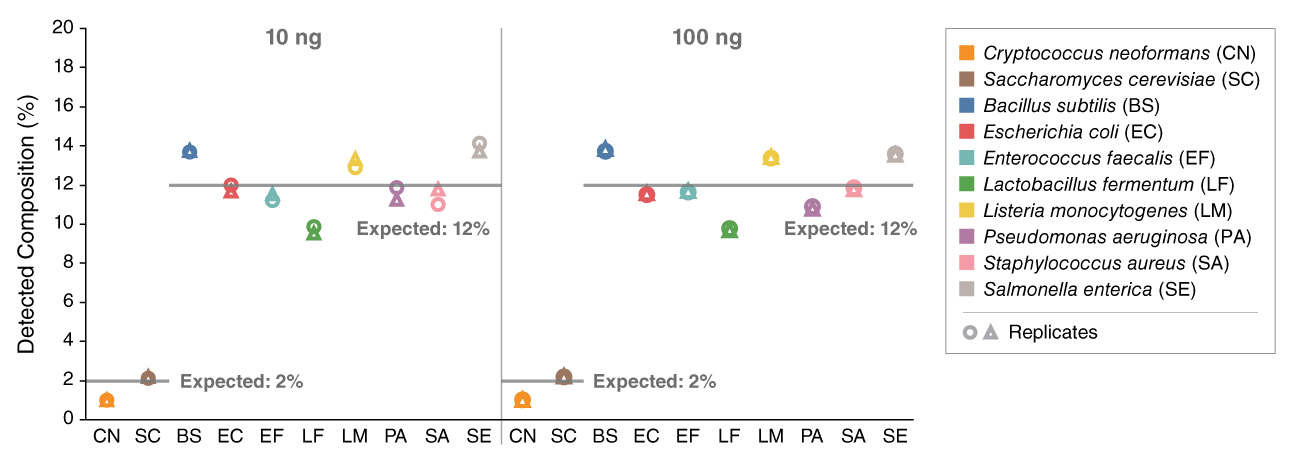
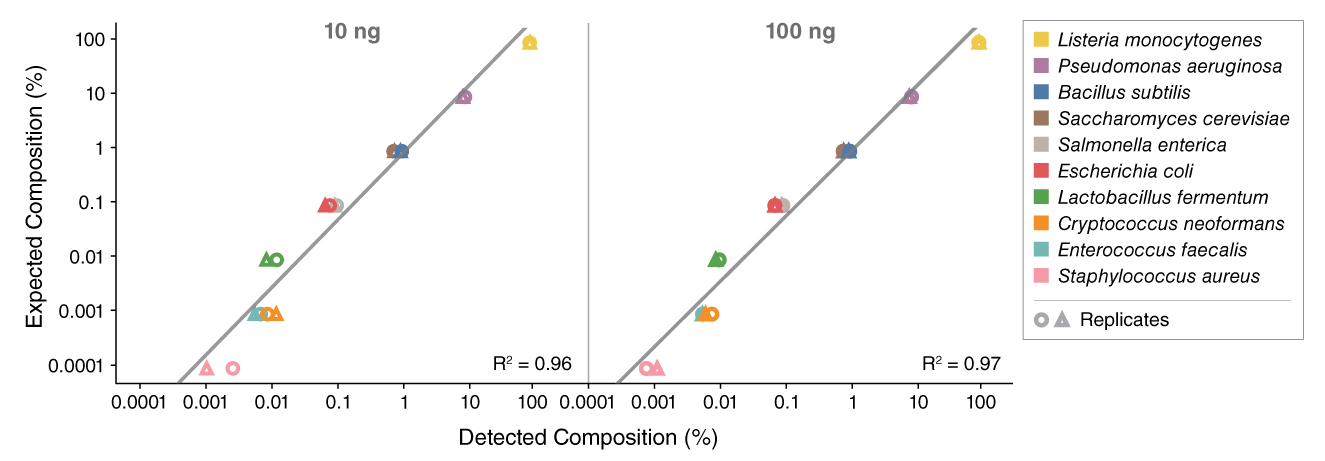
View or download extensive performance data below and in our Data Supplement.
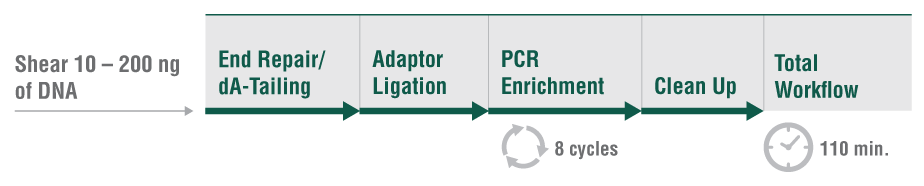
Responding to user need for a faster, streamlined DNA prep workflow that delivers high quality libraries from a variety of sample types, the NEBNext UltraExpress DNA Library Prep Kit has a single protocol for all DNA inputs (10 – 200 ng of pre-sheared DNA). The workflow incorporates master mixed reagents, reduced incubation times, and fewer cleanup steps. Use of this kit also generates less plastic consumable waste because the entire library prep is conducted in a single tube.
- Go from pre-sheared sample to library in under 2 hours
- Single-protocol simplicity cuts down on reaction setup time, while streamlined workflows speed up library prep
- A single-tube workflow means less plastic/consumable waste
- Flexibility is enabled with simple guidelines for customized protocols, if desired
- Automation-friendly protocols for enhanced scalability
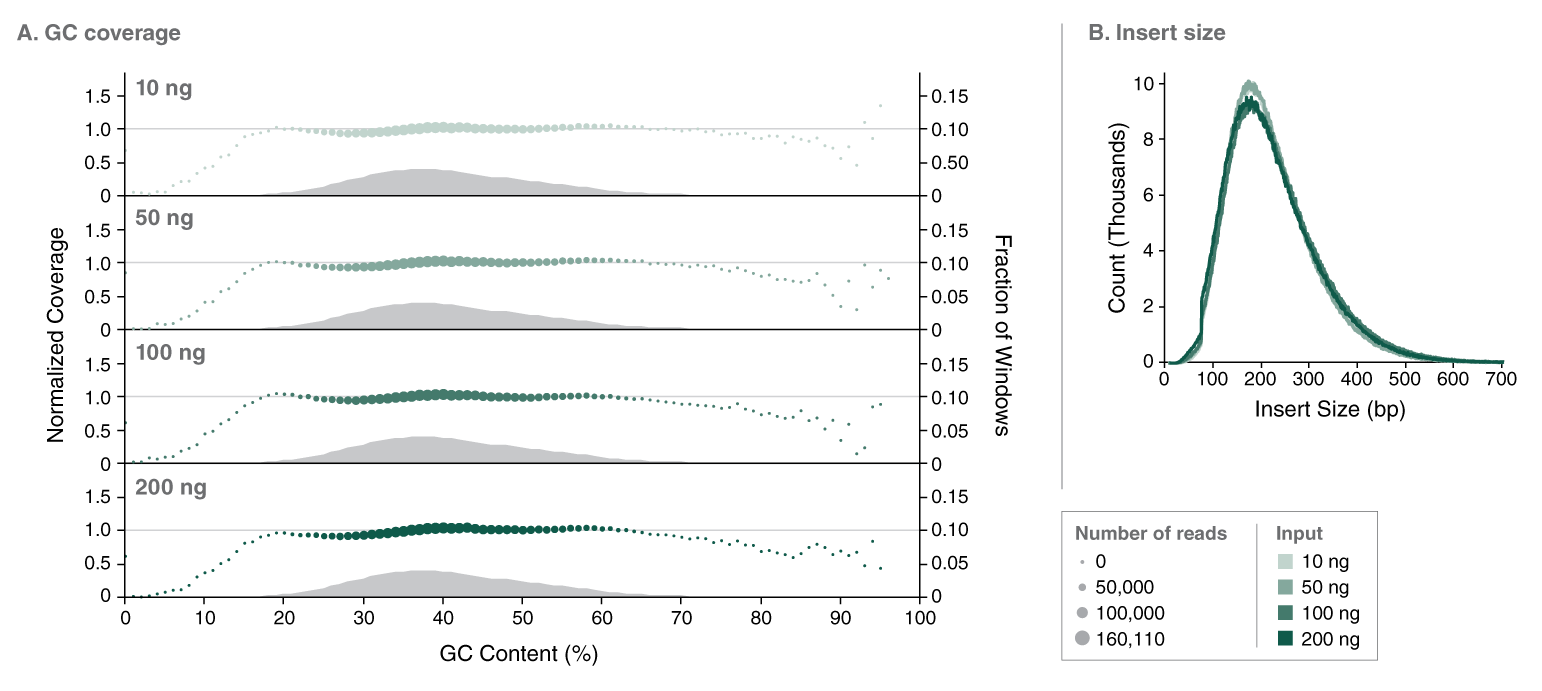
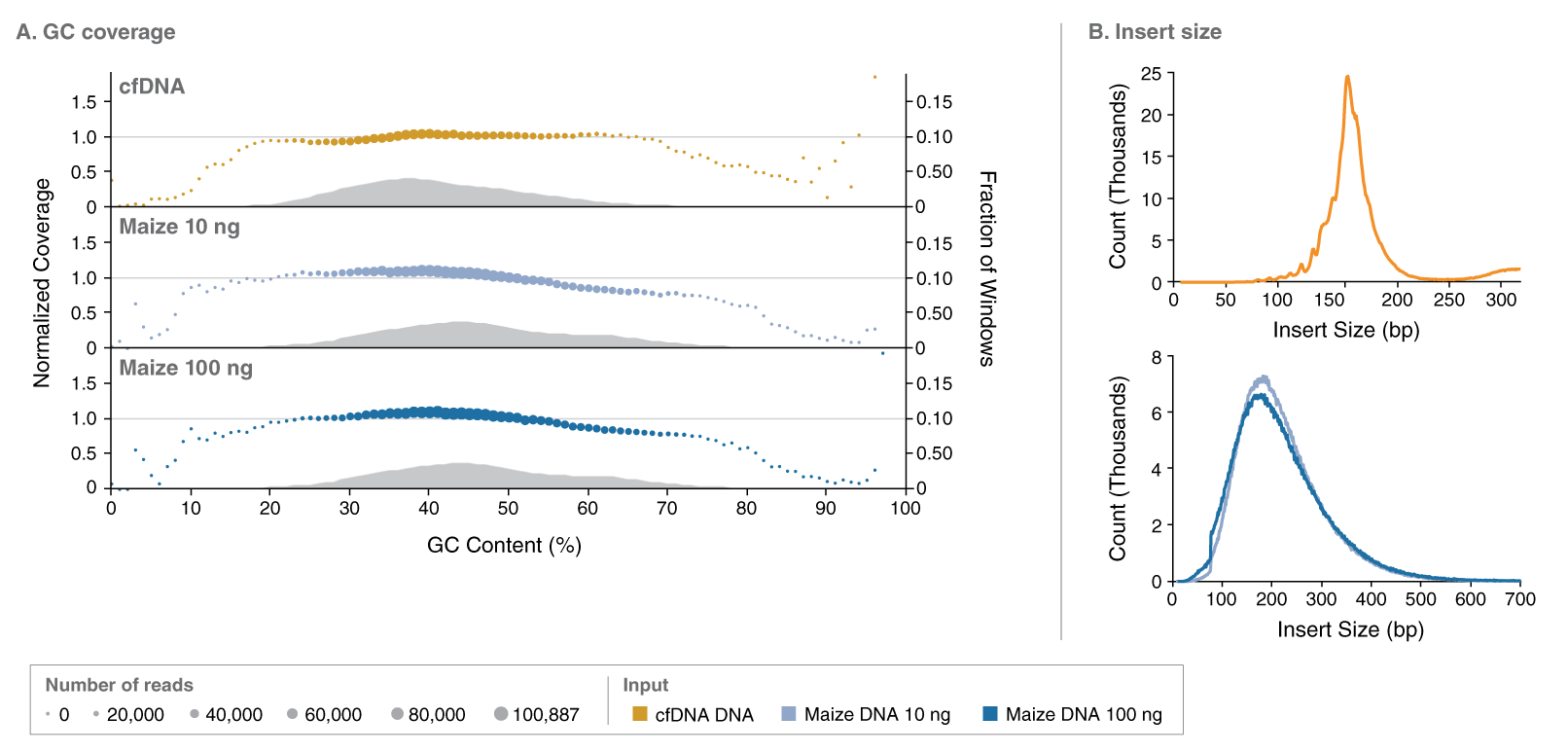
A. All sample types showed representative GC coverage. High- and low-input Maize DNA generated consistent GC coverage. The horizontal grey line indicates the expected normalized coverage of 1.0, and the colored dots represent read numbers at each GC%. The grey area plot is a histogram representing the distribution of GC content in 100 bp windows of the reference genome.
B. cfDNA insert size had the characteristic fragmentation pattern with 167 bp peak size and periodicity feature of nucleosome-bound DNA in shorter fragments. 10 ng and 100 ng Maize DNA showed consistent insert size, as expected with the shearing protocol used.
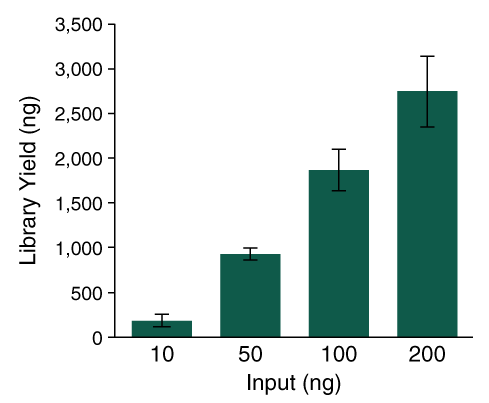
The NEBNext UltraExpress™ DNA Library Prep Kit provides robust library complexity
- 产品类别:
- Library Preparation for Illumina Products,
- DNA Library Prep for Illumina,
- Next Generation Sequencing Library Preparation Products
-
试剂盒组成
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E3325S -20 NEBNext UltraExpress™ End Prep Reaction Buffer E3324AVIAL -20 1 x 0.056 ml Not Applicable NEBNext UltraExpress™ End Prep Enzyme Mix E3326AVIAL -20 1 x 0.024 ml Not Applicable NEBNext UltraExpress™ Ligation Master Mix E3327AVIAL -20 1 x 0.24 ml Not Applicable NEBNext® MSTC™ High Yield Master Mix E3328AVIAL -20 1 x 0.96 ml Not Applicable NEBNext® Bead Reconstitution Buffer E3339AVIAL -20 1 x 0.096 ml Not Applicable 0.1X TE E3341AVIAL -20 1 x 2.4 ml Not Applicable
-
E3325L -20 NEBNext UltraExpress™ End Prep Reaction Buffer E3324AAVIAL -20 1 x 0.221 ml Not Applicable NEBNext UltraExpress™ End Prep Enzyme Mix E3326AAVIAL -20 1 x 0.096 ml Not Applicable NEBNext UltraExpress™ Ligation Master Mix E3327AAVIAL -20 1 x 0.96 ml Not Applicable NEBNext® MSTC™ High Yield Master Mix E3328AAVIAL -20 1 x 3.84 ml Not Applicable NEBNext® Bead Reconstitution Buffer E3339AAVIAL -20 1 x 3.84 ml Not Applicable 0.1X TE E3341AAVIAL -20 1 x 9.6 ml Not Applicable
-
-
相关产品
相关产品
- NEBNext® 多样本接头引物试剂盒 5(96 种 Unique 双端 Index 引物)
- NEBNext® 文库定量试剂盒(Illumina®)
- NEBNext® 磁性分离架
- Monarch® PCR & DNA 纯化试剂盒(5 μg)
操作说明、说明书 & 用法
-
操作说明
- Where can I find protocols for using the NEBNext UltraExpress™ DNA Library Prep Kit (NEB #E3325)?
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE3325
工具 & 资源
-
Web 工具
- NEBNext Selector
FAQs & 问题解决指南
-
FAQs
- What sample types can I use with the NEBNext UltraExpress™ DNA Library Prep Kit?
- Is it possible to use enzymatic fragmentation upstream of NEBNext UltraExpress™ DNA Library Prep Kit?
- What is the recommended protocol for mechanical shearing upstream of NEBNext UltraExpress™ DNA Library Prep?
- What is the difference between the NEBNext UltraExpress™ DNA Library Prep Kit and the NEBNext® Ultra™ II DNA Library Prep Kit? How do I choose the right one?
- Do the NEBNext UltraExpress™ Library Prep kits come with beads?
- Which NEBNext® Multiplex Oligos (Adaptors & Primers) can be used with the NEBNext UltraExpress Library Prep Kits?
- What should I do if I see adaptor dimer in my NEBNext UltraExpress™ libraries?
- What is the best way to quantify my NEBNext UltraExpress™ DNA libraries?
- Can this kit be used with adaptors and primers from suppliers other than NEB?
- What do I do if I see a precipitate in the Ultra II End Prep Reaction Buffer?
