上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
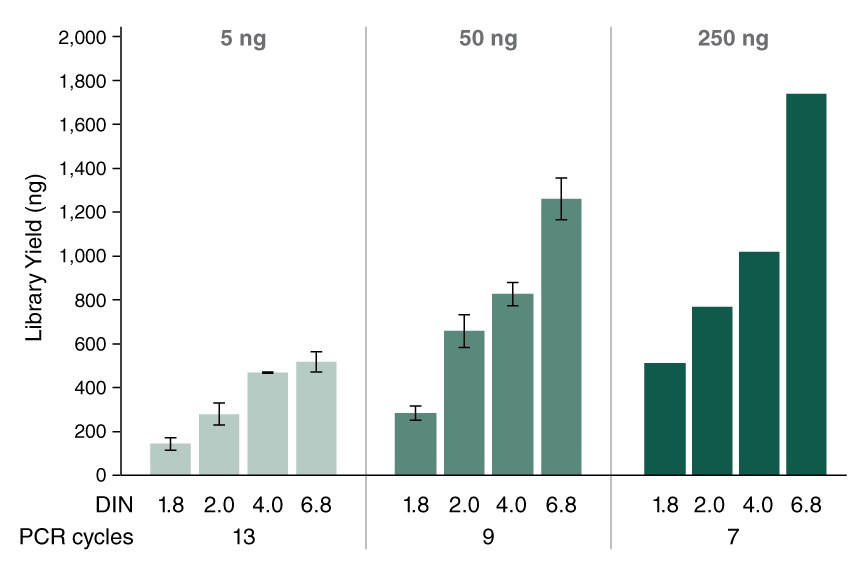
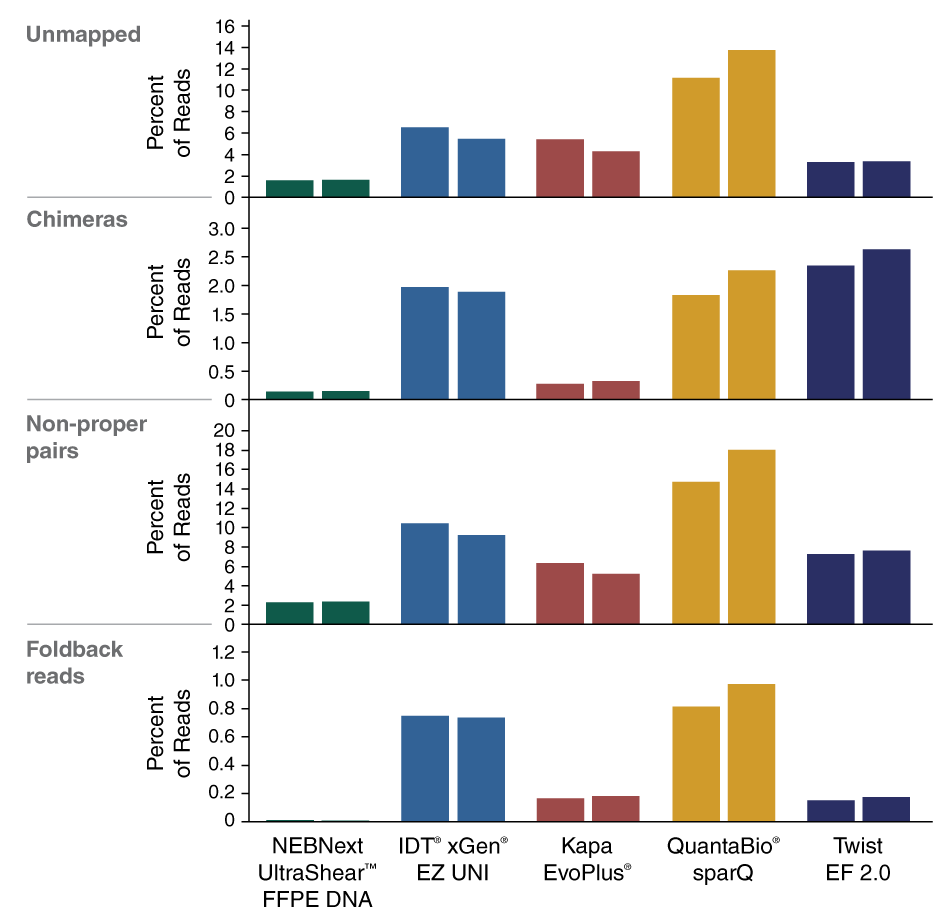
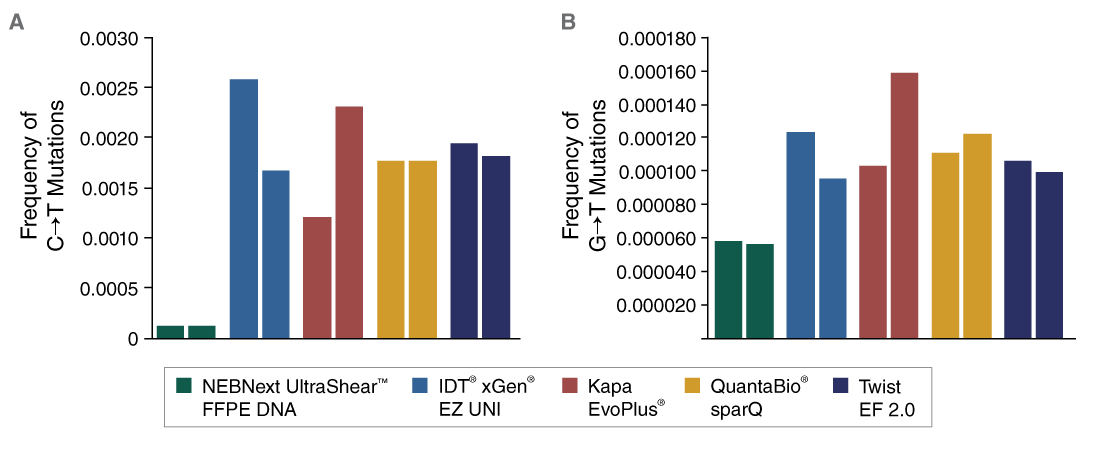
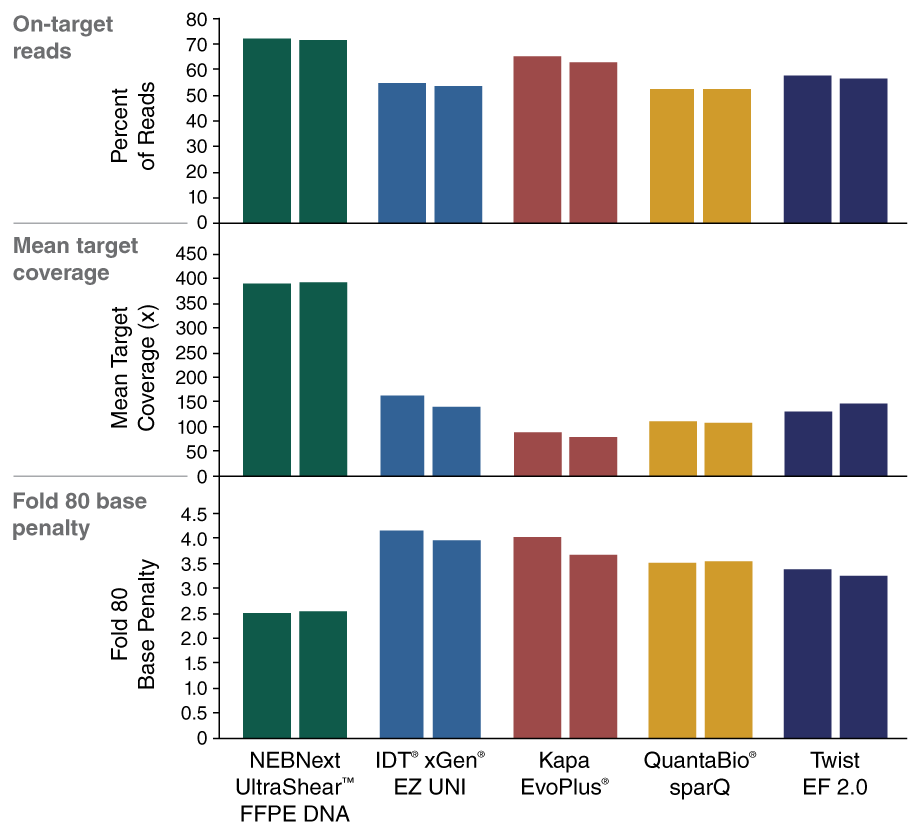
View or download extensive performance data in our Data Supplement.
FFPE DNA is often compromised in quality and quantity and is challenging for library prep and sequencing, including in target enrichment workflows that require high amounts of library input. The NEBNext UltraShear FFPE DNA Library Prep Kit addresses these challenges and improves ease of workflow and scalability:
- Repairs FFPE-induced damage using the NEBNext FFPE DNA Repair Mix v2
- Incorporates specialized enzymatic DNA fragmentation using NEBNext UltraShear
- Uses NEBNext Ultra II library prep reagents in a protocol optimized for FFPE DNA
- Robustly amplifies libraries using NEBNext MSTC™ FFPE Master Mix
- High library yields, sufficient for input into target enrichment workflows
- User-friendly workflow with minimal hands-on time and multiple safe stopping points
The NEBNext UltraShear FFPE DNA Library Prep Kit reduces damage from deamination and oxidation due to the fixation, storage, and extraction process for FFPE DNA samples, thereby reducing false positive variant calls.
The combination of specialized enzymatic FFPE DNA fragmentation and repair with optimized reagents and protocols provides superior performance with this challenging sample type.
For FFPE DNA library prep without enzymatic fragmentation, the NEBNext FFPE DNA Library Prep Kit (NEB #E6650) is available.

- 产品类别:
- FFPE DNA Products,
- Next Generation Sequencing Library Preparation Products
-
试剂盒组成
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E6655S Multi-temperature NEBNext Ultrashear® FFPE DNA Library Prep Kit E6655-2 -20 NEBNext® FFPE DNA Repair Mix v2 E7361AVIAL -20 1 x 0.048 ml Not Applicable NEBNext® Thermolabile Proteinase K E7362AVIAL -20 1 x 0.048 ml Not Applicable NEBNext UltraShear™ Reaction Buffer E6657AVIAL -20 1 x 0.168 ml Not Applicable NEBNext UltraShear™ E6656AVIAL -20 1 x 0.096 ml Not Applicable 500mM DTT E6658AVIAL -20 1 x 0.048 ml Not Applicable NEBNext® Ultra II End Prep Enzyme Mix E7646AVIAL -20 1 x 0.072 ml Not Applicable NEBNext® Ultra™ II Ligation Master Mix E7648AVIAL -20 1 x 0.72 ml Not Applicable NEBNext® Ligation Enhancer E7374AVIAL -20 1 x 0.024 ml Not Applicable NEBNext® MSTC FFPE Master Mix E6651AVIAL -20 1 x 0.6 ml 2 X NEBNext® Sample Purification Beads E6652S 25 NEBNext® Sample Purification Beads E6652AVIAL 25 1 x 4.3 ml Not Applicable
-
E6655L Multi-temperature NEBNext Ultrashear® FFPE DNA Library Prep Kit E6655-3 -20 NEBNext® FFPE DNA Repair Mix v2 E7361AAVIAL -20 1 x 0.192 ml Not Applicable NEBNext® Thermolabile Proteinase K E7362AAVIAL -20 1 x 0.192 ml Not Applicable NEBNext UltraShear™ Reaction Buffer E6657AAVIAL -20 1 x 0.672 ml Not Applicable NEBNext UltraShear™ E6656AAVIAL -20 1 x 0.384 ml Not Applicable 500mM DTT E6658AAVIAL -20 1 x 0.192 ml Not Applicable NEBNext® Ultra II End Prep Enzyme Mix E7646AAVIAL -20 1 x 0.288 ml Not Applicable NEBNext® Ultra™ II Ligation Master Mix E7648AAVIAL -20 3 x 0.96 ml Not Applicable NEBNext® Ligation Enhancer E7374AAVIAL -20 1 x 0.096 ml Not Applicable NEBNext® MSTC FFPE Master Mix E6651AAVIAL -20 2 x 1.2 ml 2 X NEBNext® Sample Purification Beads E6652L 25 NEBNext® Sample Purification Beads E6652AAVIAL 25 1 x 17 ml Not Applicable
-
-
特性和用法
需要但不提供的材料
- 80% Ethanol
- Nuclease-free Water
- 0.1X TE (1 mM Tris-HCl, pH 8.0, 0.1 mM EDTA)
- DNase-, RNase-free PCR strip tubes
- NEBNext Multiplex Oligos for Illumina® (www.neb.com/oligos)
- Magnetic rack/stand (NEB #S1515S, Alpaqua® cat. #A001322, or equivalent)
- Thermal cycler
- Agilent® Bioanalyzer® or TapeStation® and associated reagents and consumables
- Adaptor Dilution Buffer NEB #B1430S or NEBNext Unique Dual Index UMI Adaptor Dilution Buffer supplied with NEBNext Unique Dual Index UMI Adaptor DNA Sets (NEB #E7395/E7874/E7876/E7878)
-
相关产品
相关产品
- e6650nebnext-ffpe-dna-library-prep-kit
- NEBNext® 多样本接头引物试剂盒 1(96 种 Unique 双端 Index 引物)
- NEBNext® 多样本接头引物试剂盒 2(96 种 Unique 双端 Index 引物)
- NEBNext® Multiplex Oligos for Illumina® (96 Unique Dual Index Primer Pairs Set 3)
操作说明、说明书 & 用法
-
操作说明
- Where can I find guidelines and protocols for use of the NEBNext UltaShear FFPE DNA Library Prep Kit (NEB #E6655)?
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE6655
工具 & 资源
-
选择指南
- NEBNext® Multiplex Oligos Selection Chart
FAQs & 问题解决指南
-
FAQs
- What sample types can I use with the NEBNext UltraShear FFPE DNA Library Prep Kit?
- How much starting material is required for the NEBNext UltraShear FFPE DNA Library Prep Kit?
- When preparing samples using the NEBNext UltraShear FFPE DNA Library Prep Kit, can my input DNA be in EDTA-containing solutions?
- Do I really need to vortex the NEBNext UltraShear enzyme mix before use?
- What fragmentation time should be used with the NEBNext UltraShear FFPE DNA Library Prep Kit for FFPE DNA samples?
- Is size selection recommended?
- What are the safe stopping points in this protocol?
- Which NEBNext Multiplex Oligos can be used with the NEBNext UltraShear FFPE DNA Library Prep Kit?
- What input amount (nanograms) is recommended for standard versus target enrichment library prep with FFPE DNA samples?
- Can I use this NEBNext kit with adaptors and primers from vendors other than NEB?
- Will treating my DNA with the FFPE DNA Repair Mix v2 as part of the NEBNext UltraShear FFPE DNA Library Prep Kit hurt my downstream reaction?
- Does the FFPE DNA Repair v2 Mix repair DNA-protein crosslinks?
- Does the FFPE DNA Repair v2 Mix fix blocked 3′ ends?
- Can the FFPE DNA Repair v2 Mix repair damage in both single- and double-stranded DNA? Or, does it require double stranded DNA as a template?
- If I had a DNA template with mutation sites (i.e. 8-oxoguanine or deaminated cytosines) that are directly adjacent to each other on opposite strands would treatment with the FFPE DNA Repair v2 Mix cause a double strand nick/break?
- What gap lengths can be repaired with the FFPE DNA Repair v2 Mix?
