上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
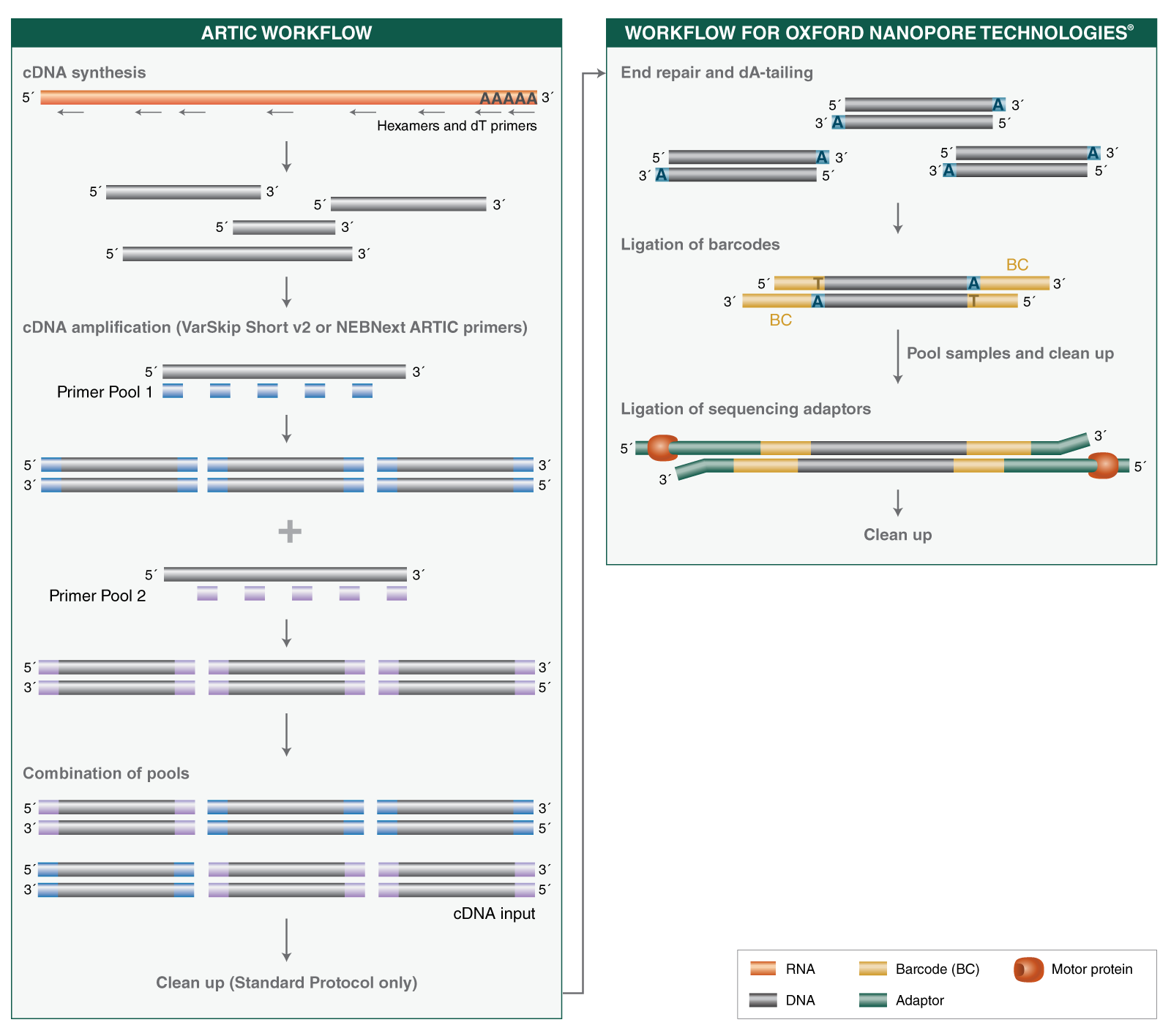
The ARTIC SARS-CoV-2 protocol from the ARTIC Network is a multiplexed amplicon approach covering the whole viral genome. The NEBNext ARTIC SARS-CoV-2 Library Prep Kit is based on this method and incorporates optimized and novel reagents.
A single RT protocol is used regardless of input amount (10-10,000 viral genome copies), and no normalization step is required ahead of targeted amplification.
This kit includes two options for primers, including new VarSkip Short v2 primers:
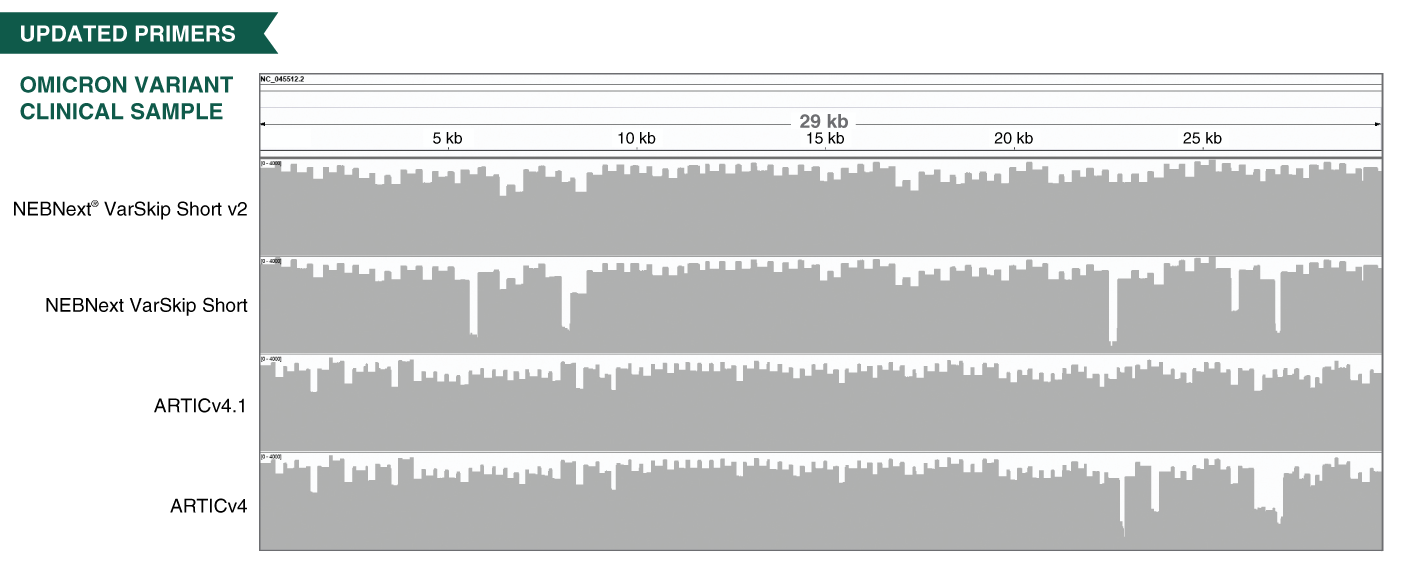
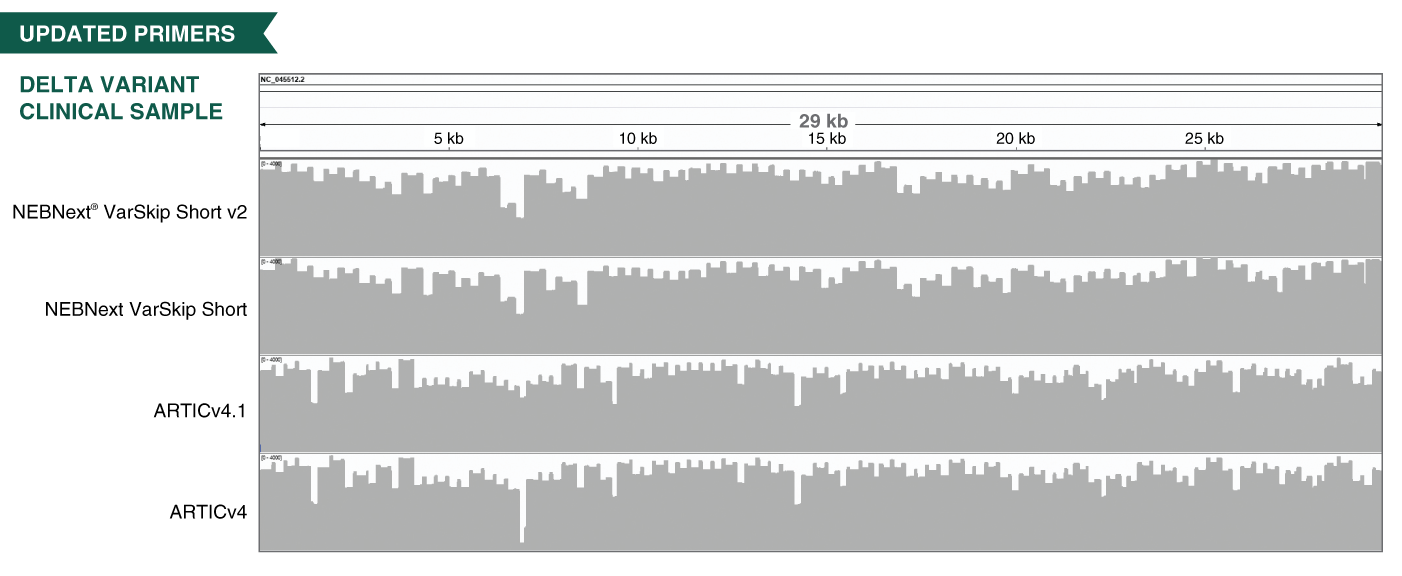
- The VarSkip (for Variant Skip) Short primers have been designed at NEB to reduce the impact of SARS-CoV-2 variants on amplification and provide improved performance, including with the Omicron variant. VarSkip Short v2 primer pools generate ~550bp amplicons. Please see the FAQ section and the product manual for additional detail. Information on primer variant overlaps can be found here.
- The V3 ARTIC primers have been balanced, using methodology developed at NEB based on empirical data from sequencing. In combination with optimized reagents for RT-PCR, the kits deliver improved uniformity of amplicon yields from gRNA across a wide copy number range.
The Express protocol removes the cleanup of cDNA Amplicons.
The NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies) is compatible with sequencing on the Oxford Nanopore Technologies platform and is used alongside kits from Oxford Nanopore Technologies to generate libraries with inserts in the 400bp range, that can be sequenced on any Oxford Nanopore Technologies instrument, in 2-24 hour run times.
If Illumina-compatible libraries are preferred, the NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit (Illumina) (NEB #E7658) and the NEBNext ARTIC SARS-CoV-2 Library Prep Kit (Illumina) (NEB #E7650) generate libraries with ~150 bp and ~400 bp inserts, respectively. If only the reagents for cDNA synthesis and targeted cDNA amplification are required, the NEBNext ARTIC SARS-CoV-2 RT-PCR Module (NEB #E7626) is available. Note that E7650 does not include VarSkip v2 primers.
- 产品类别:
- NEBNext® ARTIC products for SARS-CoV-2 sequencing,
- RNA Library Prep for Illumina,
- Next Generation Sequencing Library Preparation Products
-
试剂盒组成
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E7660S Multi-temperature NEBNext® ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies®) E7660-1 -20 LunaScript® RT SuperMix E7651AVIAL -20 1 x 0.048 ml 5 X Q5® Hot Start High-Fidelity 2X Master Mix E7652AVIAL -20 1 x 0.3 ml 2 X NEBNext® Ultra II™ End Prep Enzyme Mix E7661AVIAL -20 1 x 0.018 ml Not Applicable NEBNext® Ultra II™ End Prep Buffer E7662AVIAL -20 1 x 0.042 ml Not Applicable Blunt/TA Ligase Master Mix E7663AVIAL -20 1 x 0.24 ml Not Applicable NEBNext® Quick T4 DNA Ligase E7664AVIAL -20 1 x 0.02 ml Not Applicable NEBNext® Quick Ligation Reaction Buffer E7665AVIAL -20 1 x 0.04 ml 5 X Nuclease-free Water E7667AVIAL -20 1 x 1.5 ml Not Applicable NEBNext® Sample Purification Beads E7666S 25 NEBNext® Sample Purification Beads E7666SVIAL 25 1 x 0.872 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mixes E7871S -20 NEBNext® ARTIC SARS-CoV-2 Primer Mix 1 E7725AVIAL -20 1 x 0.042 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 2 E7726AVIAL -20 1 x 0.042 ml Not Applicable NEBNext® ARTIC Human Control Primer Pairs 1 E7727AVIAL -20 1 x 0.007 ml Not Applicable NEBNext® ARTIC Human Control Primer Pairs 2 E7728AVIAL -20 1 x 0.007 ml Not Applicable NEBNext® VarSkip Short v2 SARS-CoV-2 Primer Mix 1 E7872AVIAL -20 1 x 0.042 ml Not Applicable NEBNext® VarSkip Short v2 SARS-CoV-2 Primer Mix 2 E7873AVIAL -20 1 x 0.042 ml Not Applicable
-
E7660L Multi-temperature NEBNext® ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies®) E7660-2 -20 LunaScript® RT SuperMix E7651AAVIAL -20 1 x 0.192 ml 5 X Q5® Hot Start High-Fidelity 2X Master Mix E7652AAVIAL -20 1 x 1.2 ml 2 X NEBNext® Ultra II™ End Prep Enzyme Mix E7661AAVIAL -20 1 x 0.072 ml Not Applicable NEBNext® Ultra II™ End Prep Buffer E7662AAVIAL -20 1 x 0.168 ml Not Applicable Blunt/TA Ligase Master Mix E7663AAVIAL -20 1 x 0.96 ml Not Applicable NEBNext® Quick T4 DNA Ligase E7664AVIAL -20 1 x 0.02 ml Not Applicable NEBNext® Quick Ligation Reaction Buffer E7665AVIAL -20 1 x 0.04 ml 5 X Nuclease-free Water E7667AAVIAL -20 1 x 4.7 ml Not Applicable NEBNext® Sample Purification Beads E7666L 25 NEBNext® Sample Purification Beads E7666LVIAL 25 1 x 2.9 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mixes E7871L -20 NEBNext® ARTIC SARS-CoV-2 Primer Mix 1 E7725AAVIAL -20 1 x 0.168 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 2 E7726AAVIAL -20 1 x 0.168 ml Not Applicable NEBNext® ARTIC Human Control Primer Pairs 1 E7727AVIAL -20 1 x 0.007 ml Not Applicable NEBNext® ARTIC Human Control Primer Pairs 2 E7728AVIAL -20 1 x 0.007 ml Not Applicable NEBNext® VarSkip Short v2 SARS-CoV-2 Primer Mix 1 E7872AAVIAL -20 1 x 0.168 ml Not Applicable NEBNext® VarSkip Short v2 SARS-CoV-2 Primer Mix 2 E7873AAVIAL -20 1 x 0.168 ml Not Applicable
-
-
特性和用法
需要但不提供的材料
- 80% Ethanol (freshly prepared)
- DNA LoBind Tubes (Eppendorf #022431021)
- Oxford Nanopore Technologies Native Barcoding Expansion kits 1-12 (EXP-NBD104), 13-24 (EXP-NBD114) or 1-96 (EXP-NBD196)
- Oxford Nanopore Technologies Ligation Sequencing Kit (SQK-LSK109)
- Oxford Nanopore Technologies SFB Expansion Kit (EXP-SFB001)
- Qubit® dsDNA HS Assay Kit (ThermoFisher® Q32851)
- Magnetic rack/stand (NEB #S1515, Alpaqua®, cat. #A001322 or equivalent)
- Thermal cycler
- Vortex Mixer
- Microcentrifuge
- Agilent® Bioanalyzer® or similar fragment analyzer and associated consumables (4150 or 4200 TapeStation System)
- DNase RNase free PCR strip tubes (USA Scientific 1402-1708)
- 1.5 ml tube magnet stand (NEB #S1506)
-
优势和特性
Features
- Streamlined, high-efficiency protocol
- Effective with a wide range of viral genome inputs (10-10,000 copies)
- Improved uniformity of SARS-CoV-2 genome coverage
- Optional use control human primers provided
- Includes NEBNext Sample Purification Beads (SPRIselect®)
-
相关产品
相关产品
- E7658 NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit Illumina
- E7650 NEBNext ARTIC SARS-CoV-2 Library Prep Kit Illumina
-
参考文献
- Josh Quick (2020). nCoV-2019 sequencing protocol v2 (GunIt) . protocols.io.
操作说明、说明书 & 用法
-
操作说明
- Where can I find guidelines and protocols for using the NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies), including cDNA synthesis, targeted cDNA amplification, end repair and ligation, and the Express protocol option with reduced cleanup steps?
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE7660 with VarSkip Short SARS-CoV-2 Primers v5
- manualE7660 with VarSkip Short v2 SARS-CoV-2 Primers v6
- manualE7660 with VarSkip Short v2 SARS-CoV-2 Primers v7
工具 & 资源
-
Web 工具
- NEBNext Selector
FAQs & 问题解决指南
-
FAQs
- What is the difference between the NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes and the original NEBNext VarSkip Short SARS-CoV-2 Primer Mixes?
- What is the difference between the NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes and the NEBNext ARTIC SARS-CoV-2 Primer Mixes?
- Where can I find primer sequence information for NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes and NEBNext ARTIC SARS-CoV-2 Primer Mixes?
- Can the NEBNext ARTIC SARS-CoV-2 Primer Mixes and NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes be added to the same targeted amplification reaction?
- Which primer scheme should I use: NEBNext ARTIC SARS-CoV-2 Primer Mixes or NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes?
- Can the NEBNext VarSkip Short v2 SARS-CoV-2 Primers cover non-variant SARS-CoV-2 samples?
- Which SARS-CoV-2 variants can the NEBNext VarSkip Short v2 SARS-CoV-2 Primers cover?
- Can the NEBNext ARTIC Human Control Primer Pairs and NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes be added to the same targeted amplification reaction?
- How do I analyze VarSkip Short v2 SARS-CoV-2 sequencing data?
- Are the sequences of the primers in the NEBNext ARTIC SARS-CoV-2 Primer Mixes the same as the ARTIC Network V3 primers?
- What Ct values are recommended for inputs used with the NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies)?
- If I am using 96 samples, can I skip the cleanup step after the Targeted cDNA Amplification?
- How much data do I need when sequencing libraries generated using the NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies)?
- How do I analyze my data from sequencing libraries generated using the NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies)?
- Which kits from Oxford Nanopore Technologies do I need for the complete workflow?
- Is the NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies) compatible with the ARTIC Network nCoV-2019 sequencing protocol v3 (LoCost) or the Oxford Nanopore Technologies Eco PCR Tiling protocol?
- Are there tips to avoid barcode cross contamination?
- Is the NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies) compatible with all ONT instruments?
- Does the NEBNext ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies) kit include sufficient beads for the entire ARTIC library prep workflow?
- Are there steps I can take if the pore occupancy number is low when performing sequencing?
- Can I use less of the native barcode adaptor during the ligation step (1.5.1)?
- How can the percentage of dual-barcoded reads be increased?
- What if I don’t have enough samples to be able to generate enough library DNA for sequencing?
- For high Ct samples, how can the data throughput be increased?
- Does my total RNA sample need to be DNA-free prior to starting the ARTIC workflow?
- Can coverage be improved for the BA.2 variant?
- Is the NEBNext® ARTIC SARS-CoV-2 Companion Kit (Oxford Nanopore Technologies®) E7660 compatible with Oxford Nanopore Native Barcoding Kit V14 (SQK-NBD114)?
