上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
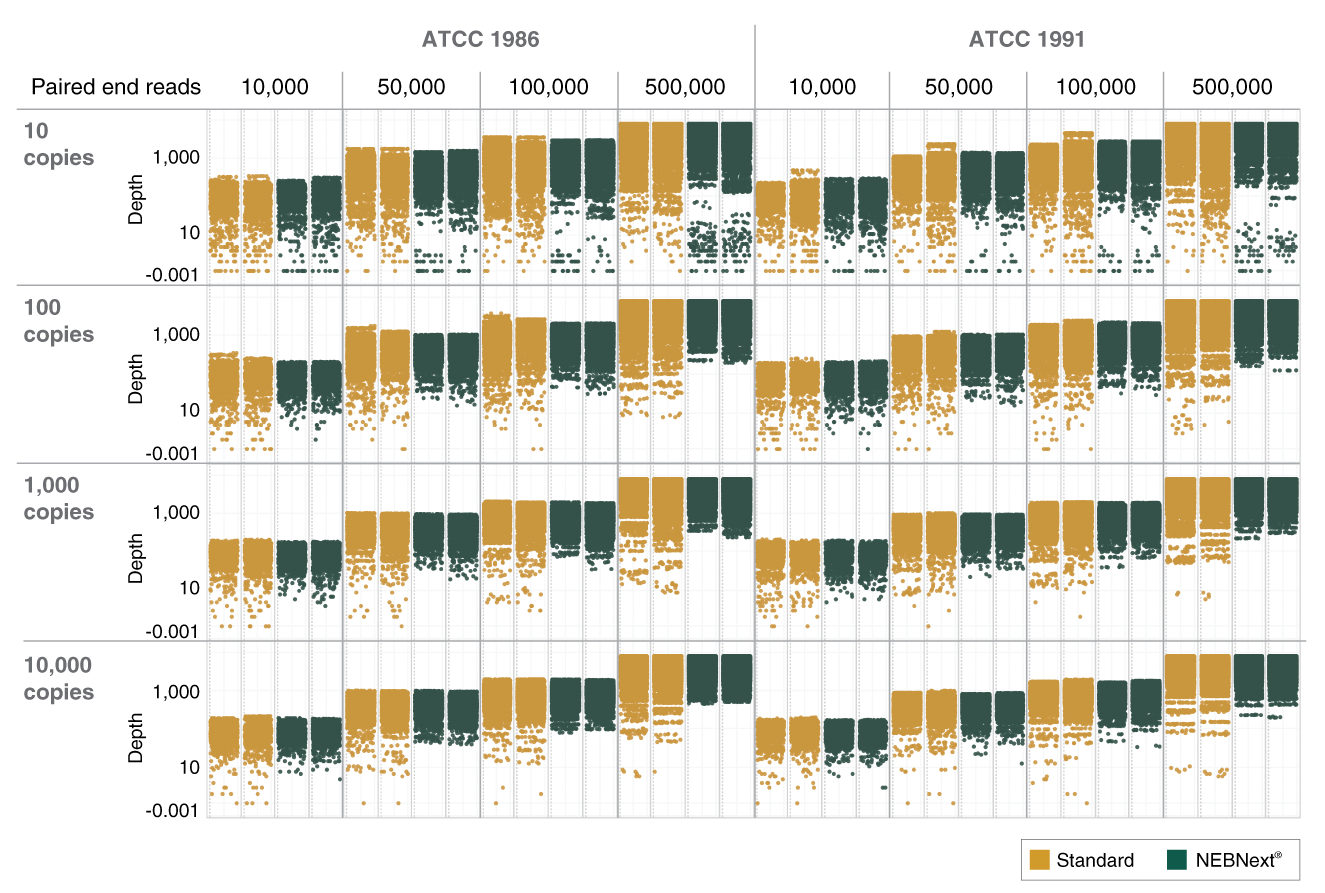
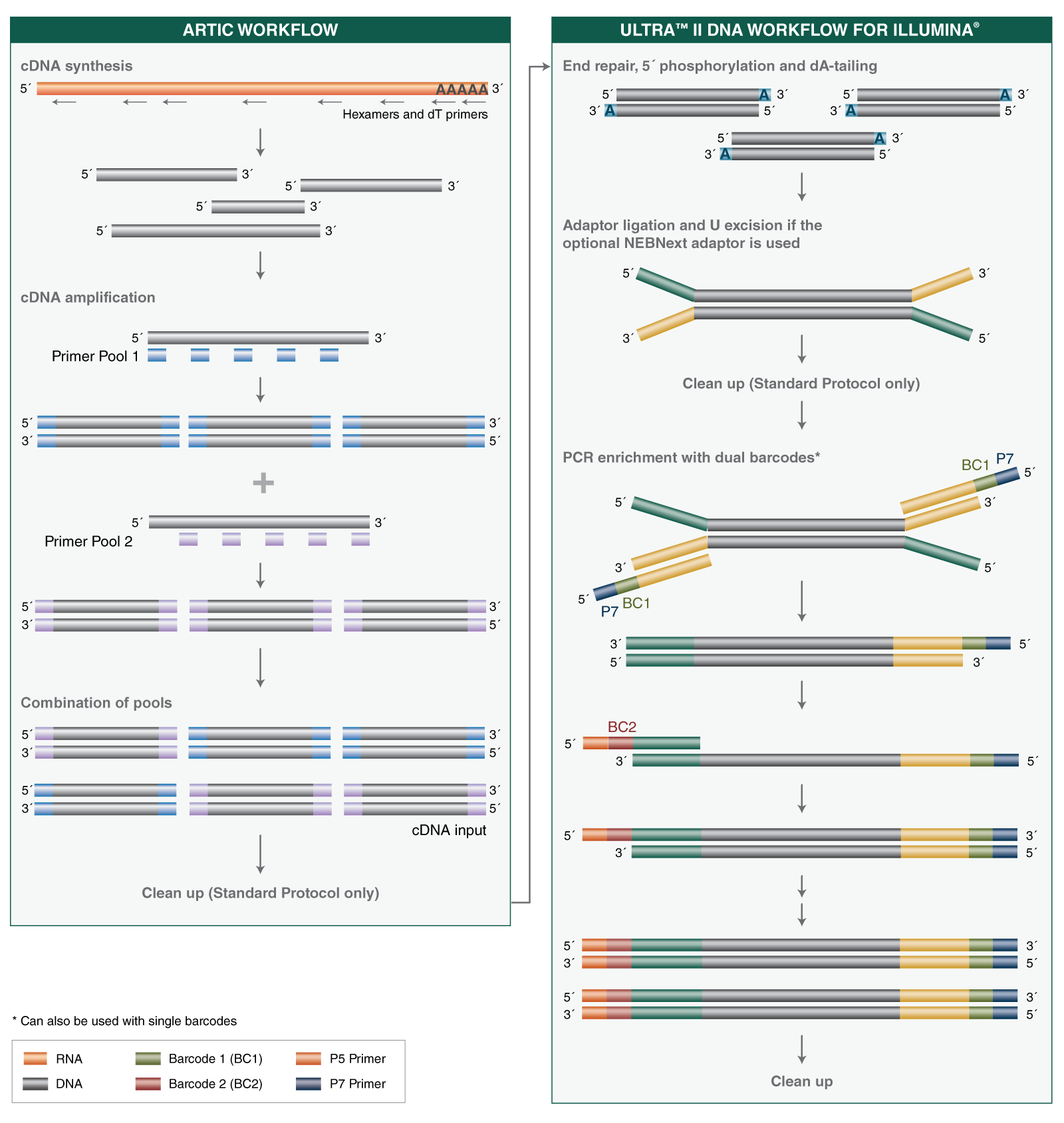
A single RT protocol is used regardless of input amount (10-10,000 viral genome copies), and no normalization step is required ahead of targeted amplification. The V3 ARTIC primers have been balanced, using methodology developed at NEB based on empirical data from sequencing, to provide uniform amplicon coverage across the viral genome. Library preparation uses the NEBNext Ultra II DNA reagents and workflow, with reagent volumes tailored to the ARTIC application. Note that adaptors and primers for library prep are purchased separately. The novel NEBNext Library PCR Master Mix allows use of the same number of PCR cycles to amplify all libraries, regardless of initial input amounts.
The NEBNext ARTIC SARS-CoV-2 Library Prep Kit generates libraries with inserts ~400 bp. If shorter inserts are preferred, the NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit (NEB #E7658) produces ~150 bp insert libraries compatible with 2 x 75 sequencing.


- 产品类别:
- NEBNext® ARTIC products for SARS-CoV-2 sequencing,
- RNA Library Prep for Illumina,
- Next Generation Sequencing Library Preparation Products
-
试剂盒组成
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E7650S Multi-temperature NEBNext® ARTIC SARS-CoV-2 Library Prep Kit (Illumina®) E7650-1 -20 LunaScript® RT SuperMix E7651AVIAL -20 1 x 0.048 ml 5 X Q5® Hot Start High-Fidelity 2X Master Mix E7652AVIAL -20 1 x 0.3 ml 2 X NEBNext® Ultra II™ End Prep Enzyme Mix E7653AVIAL -20 1 x 0.036 ml Not Applicable NEBNext® Ultra II™ End Prep Buffer E7654AVIAL -20 1 x 0.084 ml Not Applicable NEBNext® Ultra II™Ligation Master Mix E7655AVIAL -20 1 x 0.36 ml Not Applicable NEBNext® Library PCR Master Mix E7656AVIAL -20 1 x 0.3 ml 2 X 0.1X TE E7657AVIAL -20 1 x 1.3 ml Not Applicable Nuclease-free Water E7667AVIAL -20 1 x 1.5 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 1 E7725AVIAL -20 1 x 0.042 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 2 E7726AVIAL -20 1 x 0.042 ml Not Applicable NEBNext® ARTIC Human Control Primer Pairs 1 E7727AVIAL -20 1 x 0.007 ml Not Applicable NEBNext® ARTIC Human Control Primer Pairs 2 E7728AVIAL -20 1 x 0.007 ml Not Applicable NEBNext® Sample Purification Beads E7659S 25 NEBNext® Sample Purification Beads E7659SVIAL 25 1 x 2.1 ml Not Applicable
-
E7650L Multi-temperature NEBNext® ARTIC SARS-CoV-2 Library Prep Kit (Illumina®) E7650-2 -20 LunaScript® RT SuperMix E7651AAVIAL -20 1 x 0.192 ml 5 X Q5® Hot Start High-Fidelity 2X Master Mix E7652AAVIAL -20 1 x 1.2 ml 2 X NEBNext® Ultra II™ End Prep Enzyme Mix E7653AAVIAL -20 1 x 0.144 ml Not Applicable NEBNext® Ultra II™ End Prep Buffer E7654AAVIAL -20 1 x 0.336 ml Not Applicable NEBNext® Ultra II™Ligation Master Mix E7655AAVIAL -20 2 x 0.72 ml Not Applicable NEBNext® Library PCR Master Mix E7656AAVIAL -20 1 x 1.2 ml 2 X 0.1X TE E7657AAVIAL -20 1 x 5.2 ml Not Applicable Nuclease-free Water E7667AVIAL -20 1 x 1.5 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 1 E7725AAVIAL -20 1 x 0.168 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 2 E7726AAVIAL -20 1 x 0.168 ml Not Applicable NEBNext® ARTIC Human Control Primer Pairs 1 E7727AVIAL -20 1 x 0.007 ml Not Applicable NEBNext® ARTIC Human Control Primer Pairs 2 E7728AVIAL -20 1 x 0.007 ml Not Applicable NEBNext® Sample Purification Beads E7659L 25 NEBNext® Sample Purification Beads E7659SVIAL 25 4 x 2.1 ml Not Applicable
-
-
特性和用法
需要但不提供的材料
- NEBNext Singleplex or Multiplex Oligos for Illumina – www.neb.com/oligos
- 80% Ethanol (freshly prepared)
- Nuclease-free Water
- DNA LoBind Tubes (Eppendorf #022431021)
- Magnetic rack/stand (NEB #S1515, Alpaqua®, cat. #A001322 or equivalent)
- Thermocycler
- Vortex Mixer
- Microcentrifuge
- Agilent® Bioanalyzer® or similar fragment analyzer and associated consumables
- DNase RNase free PCR strip tubes (USA Scientific 1402-1708)
-
优势和特性
Features
- Streamlined, high-efficiency protocol
- Effective with a wide range of viral genome inputs (10-10,000 copies)
- Improved uniformity of SARS-CoV-2 genome coverage depth with a more balanced primer pool
- No requirement for amplicon normalization prior to library preparation
- Library inserts in the 400 bp range
- Compatible with 2 x 250 Illumina sequencing
- Includes NEBNext Sample Purification Beads (SPRIselect®)
- Adaptors and primers available separately; For a 384 unique dual index format, please contact us
-
相关产品
相关产品
- E7658 NEBNext ARTIC SARS-CoV-2 FS Library Prep Kit Illumina
- E7660 NEBNext ARTIC SARS-CoV-2 Companion Kit Oxford Nanopore Technologies
-
参考文献
- Josh Quick nCoV-2019 sequencing protocol v2 (GunIt). protocols.io.
操作说明、说明书 & 用法
-
操作说明
- Where can I find guidelines and protocols for using the NEBNext ARTIC SARS-CoV-2 Library Prep Kit, including cDNA synthesis, cDNA amplification and library prep?
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE7650
工具 & 资源
-
Web 工具
- NEBNext Selector
FAQs & 问题解决指南
-
FAQs
- Are the sequences of the primers in the NEBNext ARTIC SARS-CoV-2 Primer Mixes the same as the ARTIC Network V3 primers?
- How many sequencing reads do I need for sequencing of libraries generated using the NEBNext ARTIC Kits for Illumina?
- What Ct values are recommended for inputs used with the NEBNext ARTIC kits for Illumina?
- How do I analyze data from libraries generated using the NEBNext ARTIC kits for Illumina?
- Which NEBNext adaptors and primers are recommended for use with the ARTIC kits for Illumina?
- Do I need to add PhiX to the sequencing run?
- Do I need to normalize library concentration prior to sequencing?
- What read length is recommended for sequencing of libraries generated using the NEBNext ARTIC SARS-CoV-2 Kit (Illumina)?
- What should the final loading concentration be for MiSeq® and NextSeq® 500/550 Illumina® sequencing platforms?
- Does my total RNA sample need to be DNA-free prior to starting the ARTIC workflow?
