上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
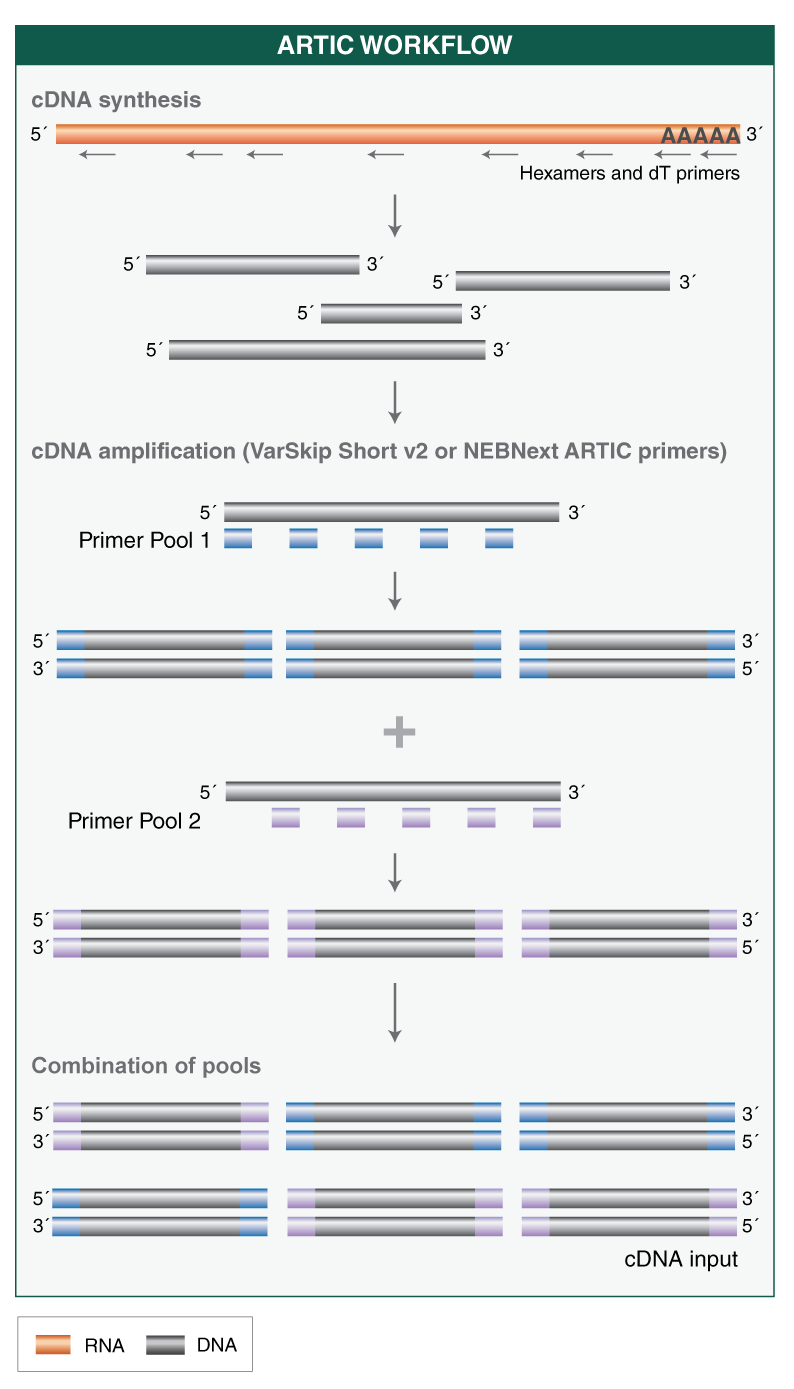
The ARTIC SARS-CoV-2 protocol from the ARTIC Network is a multiplexed amplicon approach covering the whole viral genome. The NEBNext ARTIC SARS-CoV-2 RT-PCR Module is based on this method and incorporates optimized reagents for the cDNA synthesis and targeted cDNA amplification steps.
A single RT protocol is used regardless of input amount (10-10,000 viral genome copies), and no normalization step is required ahead of targeted amplification.
This kit includes two options for primers, including new VarSkip Short v2 primers:
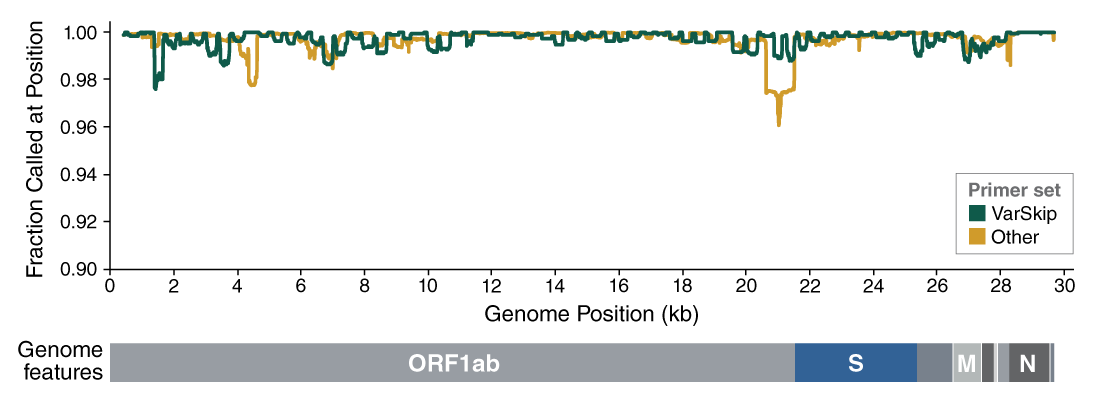
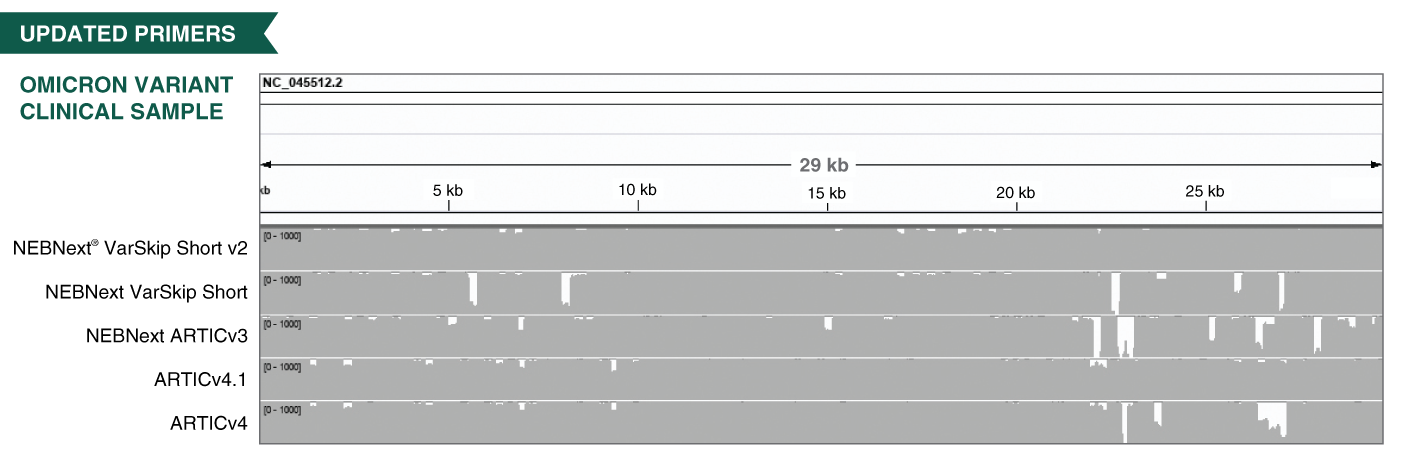
- The VarSkip (for Variant Skip) Short primers have been designed at NEB to provide improved performance with SARS-CoV-2 variants, including the Omicron variant. VarSkip Short v2 primer pools generate ~550bp amplicons. Please see the FAQ section and the product manual for additional detail. Information on primer variant overlaps can be found here.
- The V3 ARTIC primers have been balanced, using methodology developed at NEB based on empirical data from sequencing. In combination with optimized reagents for RT-PCR, the kits deliver improved uniformity of amplicon yields from gRNA across a wide copy number range.
Full kits that also include library prep reagents optimized for the ARTIC workflow are available, for smaller (~150 bp) (NEB #E7658) or larger (~400 bp) (NEB #E7650) insert libraries for Illumina® sequencing, and for Oxford Nanopore Technologies® sequencing (NEB #E7660). Note that E7650 does not include VarSkip v2 primers.
- 产品类别:
- NEBNext® ARTIC products for SARS-CoV-2 sequencing,
- RNA Library Prep for Illumina
-
产品组分信息
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
E7626S -20 LunaScript® RT SuperMix E7651AVIAL -20 1 x 0.048 ml 5 X Q5® Hot Start High-Fidelity 2X Master Mix E7652AVIAL -20 1 x 0.3 ml 2 X 0.1X TE E7657AVIAL -20 1 x 1.3 ml Not Applicable Nuclease-free Water E7667AVIAL -20 1 x 1.5 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 1 E7725AVIAL -20 1 x 0.042 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 2 E7726AVIAL -20 1 x 0.042 ml Not Applicable NEBNext® VarSkip Short v2 SARS-CoV-2 Primer Mix 1 E7872AVIAL -20 1 x 0.042 ml Not Applicable NEBNext® VarSkip Short v2 SARS-CoV-2 Primer Mix 2 E7873AVIAL -20 1 x 0.042 ml Not Applicable
-
E7626L -20 LunaScript® RT SuperMix E7651AAVIAL -20 1 x 0.192 ml 5 X Q5® Hot Start High-Fidelity 2X Master Mix E7652AAVIAL -20 1 x 1.2 ml 2 X 0.1X TE E7657AVIAL -20 2 x 1.3 ml Not Applicable Nuclease-free Water E7667AVIAL -20 1 x 1.5 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 1 E7725AAVIAL -20 1 x 0.168 ml Not Applicable NEBNext® ARTIC SARS-CoV-2 Primer Mix 2 E7726AAVIAL -20 1 x 0.168 ml Not Applicable NEBNext® VarSkip Short v2 SARS-CoV-2 Primer Mix 1 E7872AAVIAL -20 1 x 0.168 ml Not Applicable NEBNext® VarSkip Short v2 SARS-CoV-2 Primer Mix 2 E7873AAVIAL -20 1 x 0.168 ml Not Applicable
-
-
特性和用法
需要但不提供的材料
- 80% Ethanol (freshly prepared)
- Nuclease-free Water
- DNA LoBind Tubes (Eppendorf® #022431021)
- Magnetic rack/stand (NEB #S1515, Alpaqua®, cat. #A001322 or equivalent)
- Thermocycler
- Vortex Mixer
- Microcentrifuge
- Agilent® Bioanalyzer® or similar fragment analyzer and associated consumables
- DNase RNase free PCR strip tubes (USA Scientific®1402-1708)
- SPRIselect® Reagent Kit (Beckman Coulter, Inc. #B23317) or AMPure® XP Beads (Beckman Coulter, Inc. #A63881)
-
优势和特性
Features
- Improved uniformity of SARS-CoV-2 genome coverage depth with a more balanced primer pool
- Effective with a wide range of viral genome inputs (10-10,000 copies)
- Streamlined, high-efficiency protocol
-
相关产品
相关产品
- Monarch® 总 RNA 小量提取试剂盒
- NEBNext® 多样本接头引物试剂盒 1(96 种 Unique 双端 Index 引物)
- NEBNext® 多样本接头引物试剂盒 2(96 种 Unique 双端 Index 引物)
- NEBNext® Multiplex Oligos for Illumina® (96 Unique Dual Index Primer Pairs Set 3)
-
参考文献
- Josh Quick (2020). nCoV-2019 sequencing protocol v2 (GunIt). protocols.io.
操作说明、说明书 & 用法
-
操作说明
- Where can I find guidelines and protocols for using the NEBNext ARTIC SARS-CoV-2 RT-PCR Module, including cDNA synthesis and cDNA amplification?
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE7626 with VarSkip Short SARS-CoV-2 Primers v2
- manualE7626 with VarSkip Short v2 SARS-CoV-2 Primers v3
工具 & 资源
-
Web 工具
- NEBNext Selector
FAQs & 问题解决指南
-
FAQs
- What is the difference between the NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes and the original NEBNext VarSkip Short SARS-CoV-2 Primer Mixes?
- What is the difference between the NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes and the NEBNext ARTIC SARS-CoV-2 Primer Mixes?
- Where can I find primer sequence information for NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes and NEBNext ARTIC SARS-CoV-2 Primer Mixes?
- Can the NEBNext ARTIC SARS-CoV-2 Primer Mixes and NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes be added to the same targeted amplification reaction?
- Which primer scheme should I use: NEBNext ARTIC SARS-CoV-2 Primer Mixes or NEBNext VarSkip Short v2 SARS-CoV-2 Primer Mixes?
- Can the NEBNext VarSkip Short v2 SARS-CoV-2 Primers cover non-variant SARS-CoV-2 samples?
- Which SARS-CoV-2 variants can the NEBNext VarSkip Short v2 SARS-CoV-2 Primers cover?
- How do I analyze VarSkip Short v2 SARS-CoV-2 sequencing data?
- Does my total RNA sample need to be DNA-free prior to starting the ARTIC workflow?
- Can coverage be improved for the BA.2 variant?
