上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
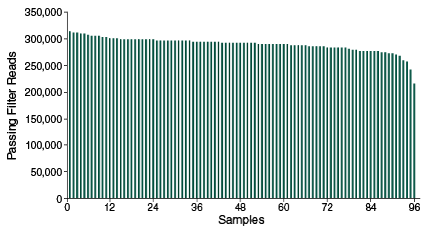
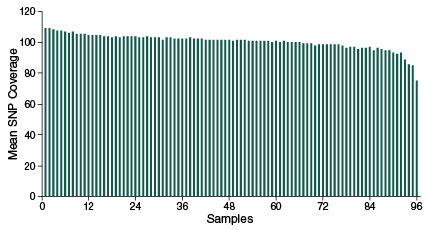
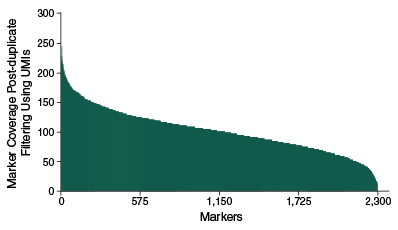
The NEBNext Direct Genotyping Solution combines highly multiplexed, capture-based enrichment with maximum efficiency next-generation sequencing to deliver cost-effective, high-throughput, genotyping for a wide variety of applications. Applicable for marker ranges spanning 100-5,000 markers, pre-capture multiplexing of up to 96 samples combined with dual indexed sequencing allows over 3.8 million genotypes in a single Illumina® sequencing run.
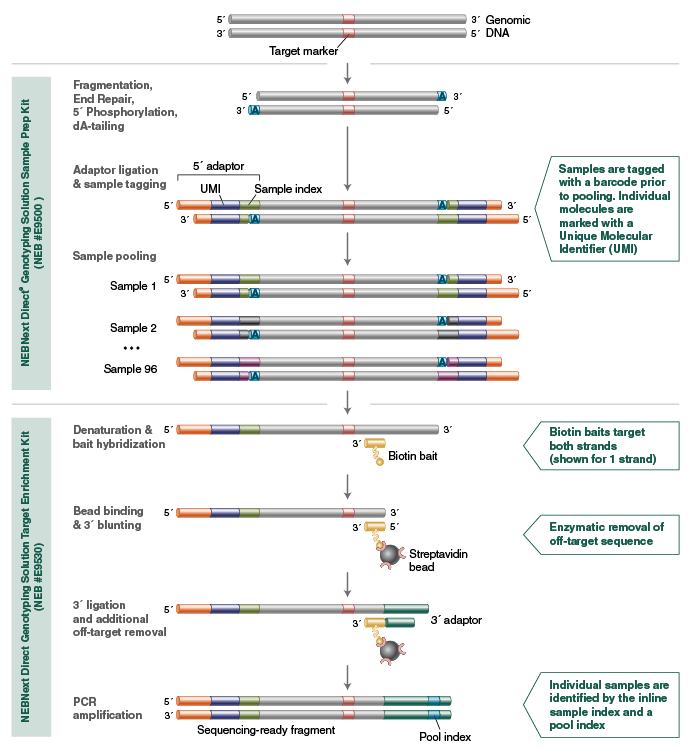
Reagents Supplied for E9500
The following reagents are supplied with this product:
| Store at (°C) | Concentration | |
| NEBNext Direct® GS Fragmentation Buffer | -20 | |
| NEBNext Direct® GS Fragmentation Enzyme Mix | -20 | |
| NEBNext Direct® GS Indexed 5′ Adaptor Plate | -20 | |
|
NEBNext Direct® GS 5′ Adaptor Ligation Master Mix |
-20 | |
| NEBNext Direct® GS Stop Solution | 4 | |
| NEBNext Direct® GS Sample Purification Beads | RT |
Reagents Supplied for E9530
The following reagents are supplied with this product:
| Store at (°C) | Concentration | |
| NEBNext Direct® GS Baits | -20 (pouch) | |
| NEBNext Direct® GS Hybridization Additive | -20 | |
| NEBNext Direct® GS 3′ Blunting Enzyme | -20 | |
| NEBNext Direct® GS 3′ Adaptor | -20 | |
| NEBNext Direct® GS Ligase | -20 | |
| NEBNext Direct® GS Specificity Enhancer Enzyme Mix | -20 | |
| NEBNext Direct® GS Q5 Master Mix | -20 | |
| NEBNext Direct® GS Index Primer Mix DGS01 | -20 | |
| NEBNext Direct® GS Index Primer Mix DGS02 | -20 | |
| NEBNext Direct® GS Index Primer Mix DGS03 | -20 | |
| NEBNext Direct® GS Index Primer Mix DGS04 | -20 | |
| NEBNext Direct® GS Index Primer Mix DGS05 | -20 | |
| NEBNext Direct® GS Index Primer Mix DGS06 | -20 | |
| NEBNext Direct® GS Index Primer Mix DGS07 | -20 | |
| NEBNext Direct® GS Index Primer Mix DGS08 | -20 | |
| NEBNext Direct® GS Pre-Hyb Sample Purification Beads | RT | |
| NEBNext Direct® GS Bead Wash 1 (BW1) | RT | |
| NEBNext Direct® GS Post-PCR Sample Purification Beads | RT | |
| NEBNext Direct® GS TE Buffer | 4 | |
| NEBNext Direct® GS Hybridization Buffer | 4 | |
| NEBNext Direct® GS Streptavidin Beads | 4 | |
| NEBNext Direct® GS Bead Prep Buffer | 4 | |
| NEBNext Direct® GS Hybridization Wash (HW) | 4 | |
| NEBNext Direct® GS Bead Wash 2 (BW2) | 4 | |
| NEBNext Direct® GS 3′ Blunting Buffer | 4 | |
| NEBNext Direct® GS 3′ Adaptor Ligation Buffer | 4 |
- 产品类别:
- Automation for NEBNext® NGS Library Prep Products
-
优势和特性
应用特性
- The most efficient targeted genotyping-by-sequencing solution available
- Pre-capture multiplexing of up to 96 samples per enrichment
- Flexibility from 100’s to 1,000’s of markers per sample
- Dual indexing plus Unique Molecular Identifier (UMI)
- Optimized bait design captures both DNA strands
- Unique and rapid, capture-based enrichment for optimal sequencing performance
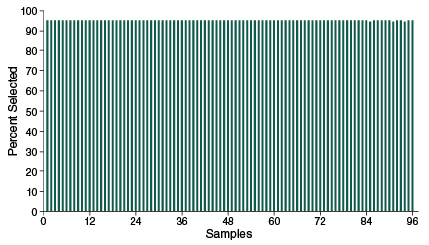
- Highly specific enrichment, > 90% of reads mapping to targets
- Coverage uniformity – sequence less and avoid over- and under-representation
- Similar performance from DNA across CTAB and column-based extraction procedures
- Fast and streamlined, automation-friendly workflow, with minimal handling steps and hands-on time’
- Pooled capture eliminates redundancy
- Single-day workflow
- Automation friendly
- The most efficient targeted genotyping-by-sequencing solution available
操作说明、说明书 & 用法
-
操作说明
- Protocol for use with NEBNext Direct Genotyping Solution (NEB #E9500 and #E9530)
- Guidelines for Using the Single-use, Indexed 5′ Adaptor Plate (NEB #E9500 and #E9530)
- Index Adaptor and Index Primer Pooling Guidelines (NEB #E9500 and #E9530)
- Guidelines for Running Samples on the Illumina MiSeq (NEB #E9500 and #E9530)
- Guidelines for Bioinformatic Processing of Sequencing Data (NEB #E9500 and #E9530)
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualE9500_E9530
-
使用指南
- bcl2fastq – Guildelines for E9500 and E9530
- bcl2fastq – Sample Sheet for E9530-768
- NEBNext Direct GS Barcodes
- Picard – Guidelines for Sample Metadata for E9500 and E9530
- Picard – Sample Metadata for E9530
- Sample Sheet – E9530 – E9530
- Sample Sheet – Guidelines for E9500B and E9530B
-
应用实例
- Fully Automated NEBNext® Direct Genotyping Solution using Beckman Coulter® Life Sciences Biomek i7 Workstation
FAQs & 问题解决指南
-
FAQs
- How many times can the -20°C reagents supplied in the NEBNext Direct® Genotyping Solution be frozen and thawed?
- How many times can the 4°C reagents be brought to room temperature?
- My NEBNext Direct Genolytyping Solution 4°C box accidentally froze, can I still use it?
- I accidentally stored the Sample Purification Beads and Bead Wash 1 (BW1) at 4°C. Can I still use them?
- What is the shelf life of the NEBNext Direct Genotyping Solution?
- What method do you recommend for extracting genomic DNA and FFPE DNA before use with the NEBNext Direct Genotyping Solution?
- What is the recommended method for quantitating my input DNA?
- If my input amount is low, should I dilute the adaptors?
- What type and how much starting material do I need to use when preparing libraries using the NEBNext Direct Genotyping Solution?
- Which magnets (2 ml and 200 µl sizes) do you recommend?
- I used Bead Wash 1 (BW1) when I should have used Bead Wash 2 (BW2) during the Post-reaction Wash. Can I do another wash with Bead Wash 2 (BW2) to save my sample?
- I used Bead Wash 2 (BW2) when I should have used Bead Wash 1 (BW1) during the Post-reaction Wash. What should I do?
- Does incomplete removal of Bead Wash 1 prior to adding Bead Wash 2 inhibit the next step?
- Can I use any Q5® High-Fidelity DNA Polymerase formulation with this kit?
- If I have adaptor dimer in my finished library, can I perform another bead cleanup to remove it?
-
How many/ few samples can I pool together for my sequencing run?
- Are blocking oligos required for this kit?
- Is the NEBNext Direct Genotyping Solution available in bulk or customized format?
