上海金畔生物科技有限公司代理New England Biolabs(NEB)酶试剂全线产品,欢迎访问官网了解更多产品信息和订购。
产品信息
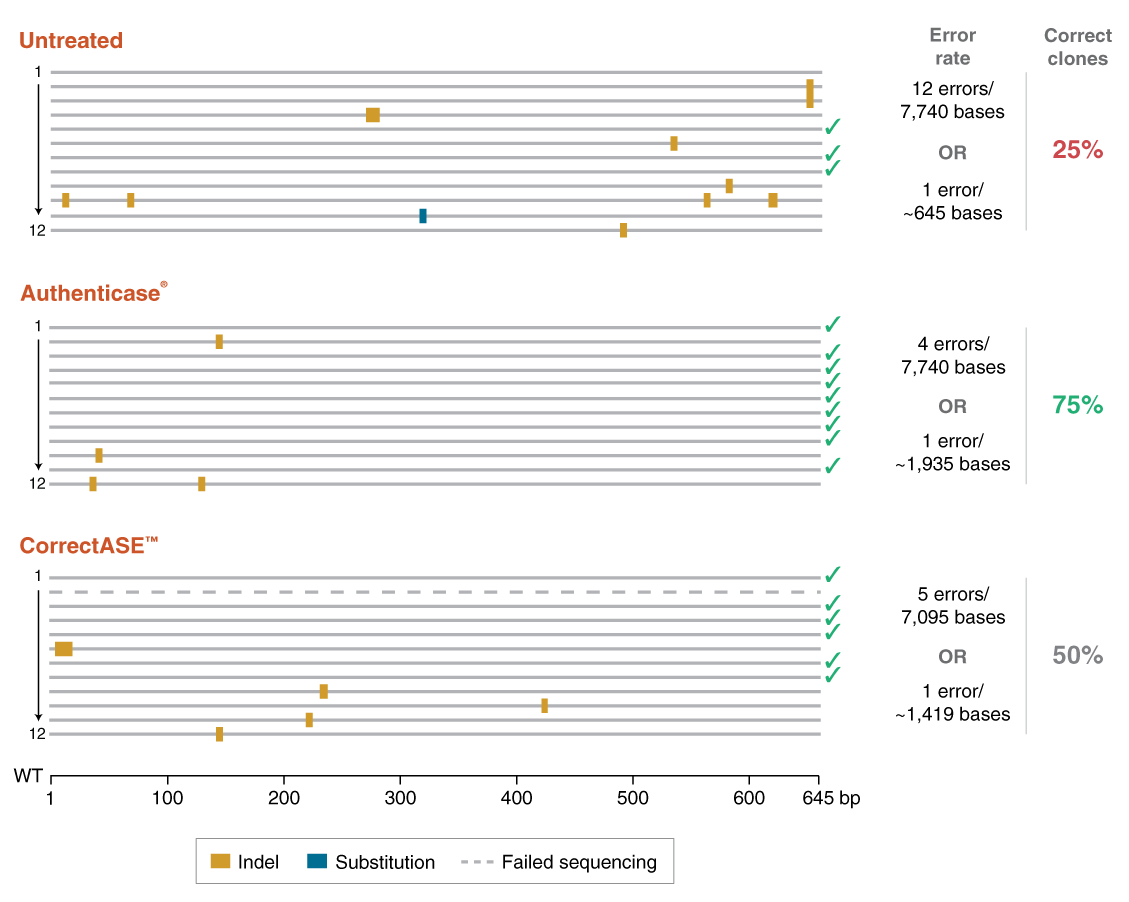
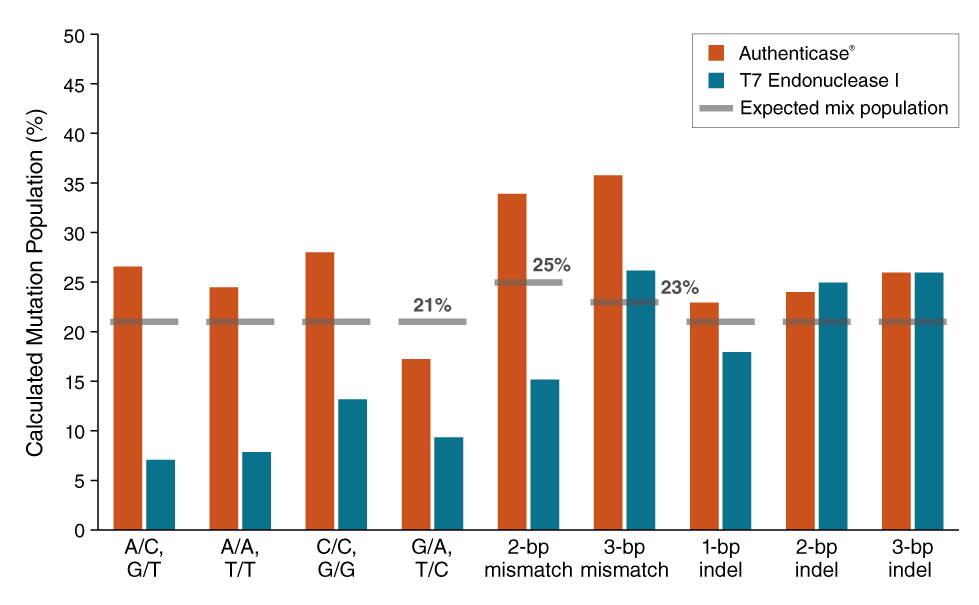
Authenticase is a proprietary mixture of structure-specific nucleases capable of recognizing and cleaving outside mismatch and indel (insertion and/or deletion) regions, ranging from 1-10 basepairs (bp) on double-stranded DNA. The formulation has limited non-specific activity on homoduplex regions of DNA. Authenticase can be used as an error-correction reagent in oligo-based PCR gene assembly by enzymatically removing “mistakes” prior to the final renaturation and amplification step (i.e. removes mismatch/indel errors caused by oligonucleotide synthesis). Alternatively, Authenticase can replace T7 Endonuclease I in the mismatch detection assay used to assess the efficiency of genome editing.

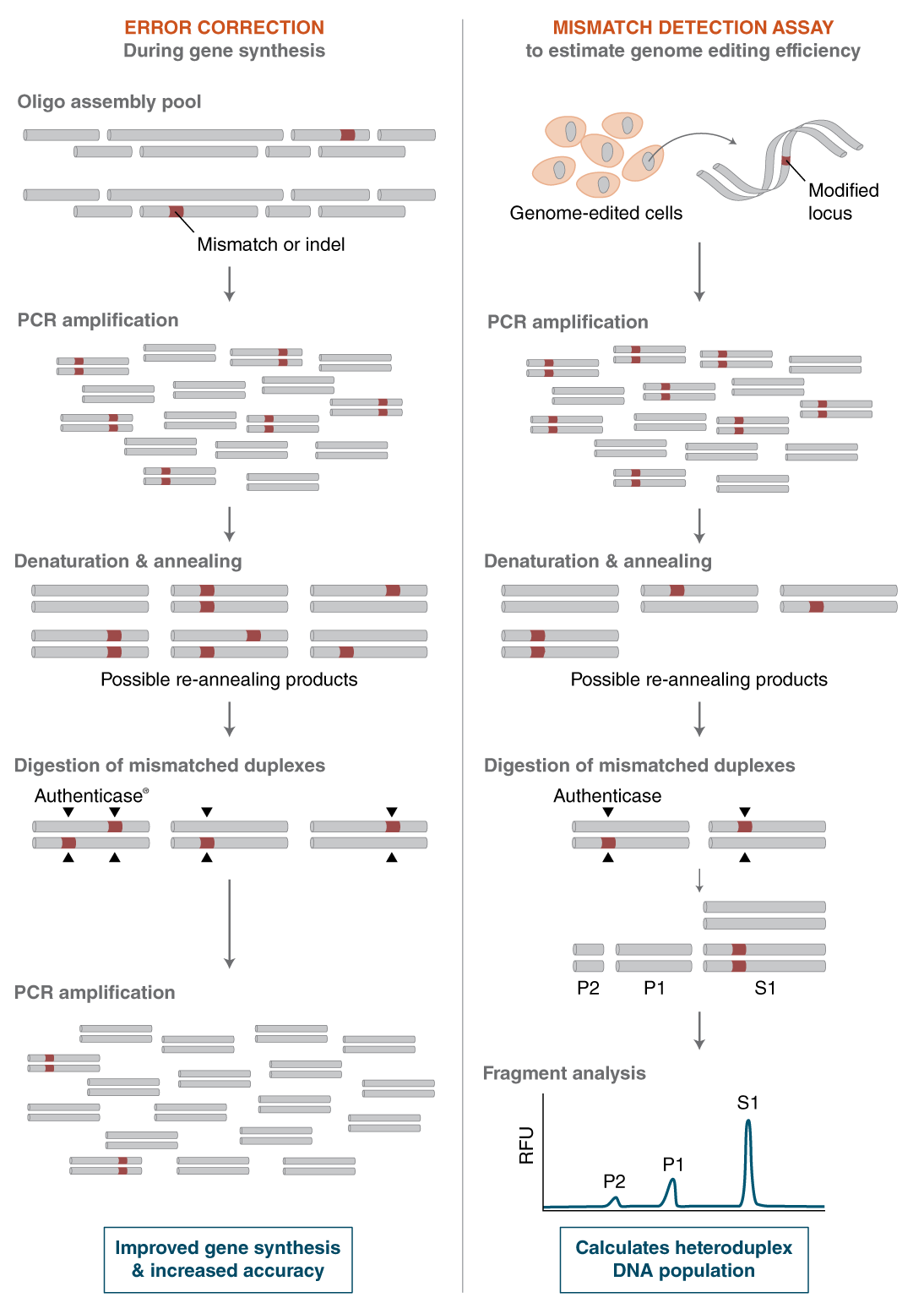
- 产品类别:
- DNA Repair Enzymes and Structure-specific Endonucleases Products
- 应用:
- Genome Editing Applications
-
产品组分信息
本产品提供以下试剂或组分:
NEB # 名称 组分货号 储存温度 数量 浓度 -
M0689S -20 Authenticase® M0689SVIAL -20 1 x 0.025 ml 25 reactions Authenticase DNA Annealing Buffer B2831SVIAL -20 1 x 0.7 ml 5 X Authenticase Reaction Buffer B2832SVIAL -20 1 x 0.5 ml 10 X
-
M0689L -20 Authenticase® M0689LVIAL -20 1 x 0.125 ml 125 reactions Authenticase DNA Annealing Buffer B2831SVIAL -20 1 x 0.7 ml 5 X Authenticase Reaction Buffer B2832SVIAL -20 1 x 0.5 ml 10 X
-
-
特性和用法
反应条件
1X Authenticase Reaction Buffer
Incubate at 42°C1X Authenticase Reaction Buffer
10 mM Tris-HCl
10 mM MgCl2
100 µg/ml Recombinant Albumin
(pH 8 @ 25°C)贮存溶液
10 mM Tris-HCl
500 mM NaCl
1 mM Dithiothreitol
0.1 mM EDTA
50% Glycerol
pH 7.4 @ 25°C热失活
否
-
优势和特性
Features
- Reduces number of colonies that need to be screened and saves time
- Can replace T7 Endonuclease I in mismatch detection assay
-
相关产品
相关产品
- Q5® 热启动超保真 2X 预混液
- M0568 Thermolabile Exonuclease I
- 快速 CIP
- Monarch® PCR & DNA 纯化试剂盒(5 μg)
- PCR Marker
- OneTaq® Quick-Load® 2X 预混液(提供标准缓冲液)
- Nuclease-free Water
- NEBuilder® 高保真 DNA 组装克隆试剂盒
操作说明、说明书 & 用法
-
操作说明
- Error Correction During Gene Synthesis (NEB #M0689)
- Mismatch Detection Assay (NEB #M0689)
- Supplemental Protocol 1: Generation of DNA fragments by PCR assembly of pooled oligos (NEB #M0689)
- Supplemental Protocol 2: Using colony PCR to identify positive clones (NEB #M0689)
-
说明书
产品说明书包含产品使用的详细信息、产品配方和质控分析。- manualM0689
工具 & 资源
-
选择指南
- Activities of DNA Repair Enzymes and Structure-specific Endonucleases
- Properties of DNA Repair Enzymes and Structure-specific Endonucleases
FAQs & 问题解决指南
-
FAQs
- What reaction conditions were used to define Authenticase™?
- How does Authenticase™ improve the quality and fidelity of PCR gene assembly?
- How do I convert my gene of interest into oligonucleotides?
- Why do you recommend setting up two tubes for the PCR reaction containing different amounts of Authenticase™-treated samples as templates?
- Can I use Authenticase™ for genome editing applications?
- Does Authenticase™ recognize single base pair mismatches or indels (insertions/deletions)?
- What common additives inhibit Authenticase™?
- What PCR reagents are recommended for DNA amplification in genome editing (CRISPR/Cas9, TALEN, ZFN) mismatch detection assays?
- Can I use Authenticase™ genome editing (CRISPR/Cas9, TALEN, ZFN) mismatch detection assays with unpurified PCR products?
- What size PCR amplicon should I design to analyze the genomic editing efficiency?
- If my PCR reaction yield is low, can I add more than 5 µl of the PCR reaction to the digestion reaction?
- Why do I see an extra band when I run the undigested heteroduplex on a Bioanalyzer or agarose gel?
- What are the differences between Mismatch Endonuclease I (NEB #M0678) and Authenticase (NEB #M0689)?
